
Currently, many applications such as Google News use clustering algorithms as the main implementation method. They can use large amounts of unlabeled data to build powerful topic clusters. This article introduces 6 types of mainstream methods from the most basic K-means clustering to powerful density-based methods. They each have their own areas of expertise and scenarios, and the basic ideas are not necessarily limited to clustering methods.

This article will start with simple and efficient K-means clustering, and then introduce mean shift clustering, density-based clustering, Gaussian mixture and maximum expectation method clustering, Hierarchical clustering and graph group detection applied to structured data. We will not only analyze the basic implementation concepts, but also give the advantages and disadvantages of each algorithm to clarify actual application scenarios.
Clustering is a machine learning technique that involves grouping data points. Given a set of data points, we can use a clustering algorithm to classify each data point into a specific group. In theory, data points belonging to the same group should have similar properties and/or characteristics, while data points belonging to different groups should have very different properties and/or characteristics. Clustering is an unsupervised learning method and a statistical data analysis technique commonly used in many fields.
K-Means is probably the most well-known clustering algorithm. It is part of many introductory data science and machine learning courses. Very easy to understand and implement in code! Please see the picture below.
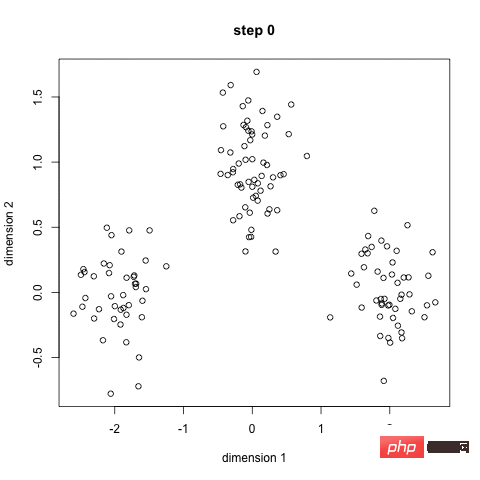
K-Means Clustering
First, we select some classes/groups and randomly initialize their respective center points. In order to work out the number of classes to use, it's a good idea to take a quick look at the data and try to identify the different groups. The center point is the location with the same length as each data point vector, which is "X" in the above image.
Classify each point by calculating the distance between the data point and the center of each group, and then classify the point in the group to which the group center is closest.
Based on these classification points, we use the mean of all vectors in the group to recalculate the group center.
Repeat these steps for a certain number of iterations, or until the group center changes little after each iteration. You could also choose to randomly initialize the group center a few times and then choose the run that seems to provide the best results.
The advantage of K-Means is that it is fast, since all we are really doing is calculating the distance between the point and the center of the group: very few calculations! So it has linear complexity O(n).
On the other hand, K-Means has some disadvantages. First, you have to choose how many groups/classes there are. This is not always done carefully, and ideally we want the clustering algorithm to help us solve the problem of how many classes to classify, since its purpose is to gain some insights from the data. K-means also starts from randomly selected cluster centers, so it may produce different clustering results in different algorithms. Therefore, results may not be reproducible and lack consistency. Other clustering methods are more consistent.
K-Medians is another clustering algorithm related to K-Means, except that instead of using the mean, the group centers are recalculated using the median vector of the group. This method is insensitive to outliers (because the median is used), but is much slower for larger data sets because a sorting is required on each iteration when calculating the median vector.
Mean Shift Clustering is a sliding window-based algorithm that attempts to find dense areas of data points. This is a centroid-based algorithm, which means that its goal is to locate the center point of each group/class, which is accomplished by updating the candidate points of the center point to the mean of the points within the sliding window. These candidate windows are then filtered in a post-processing stage to eliminate near-duplications, forming the final set of center points and their corresponding groups. Please see the legend below.
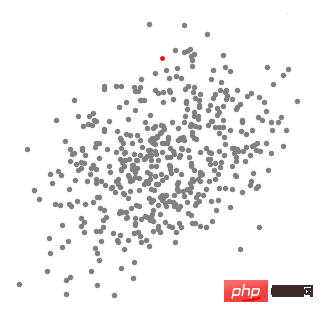
Mean shift clustering for a single sliding window
The entire process from start to finish for all sliding windows is shown below. Each black dot represents the centroid of the sliding window, and each gray dot represents a data point.
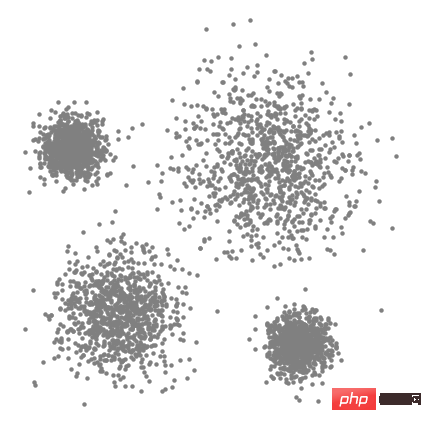
The whole process of mean shift clustering
Compared with K-means clustering, this method does not need to select the number of clusters because the mean shift automatically Discover this. This is a huge advantage. The fact that the cluster centers cluster towards the maximum point density is also very satisfying, as it is very intuitive to understand and adapt to the natural data-driven implications. The disadvantage is that the choice of window size/radius "r" may be unimportant.
DBSCAN is a density-based clustering algorithm that is similar to mean shift but has some significant advantages. Check out another fun graphic below and let’s get started!
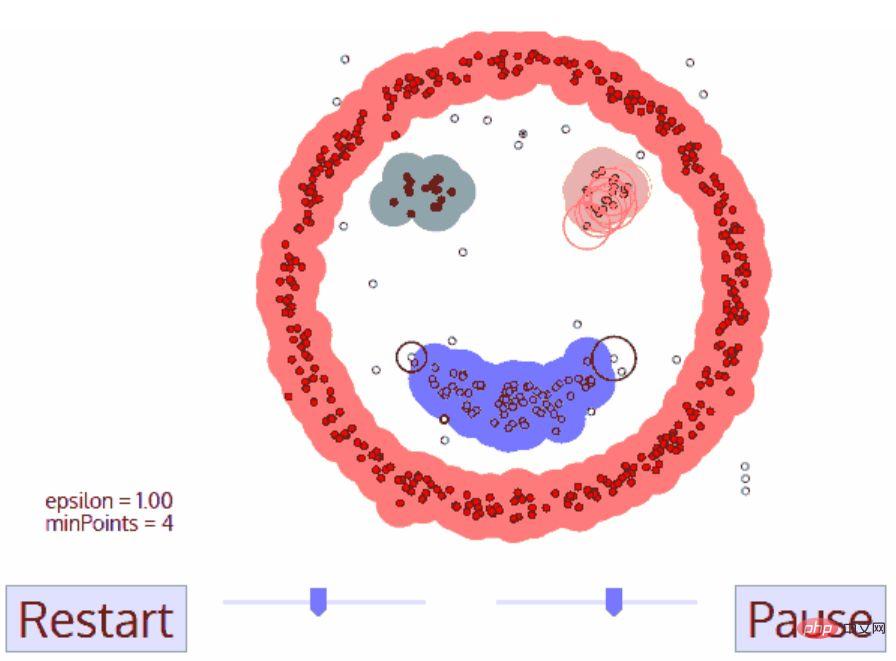
DBSCAN has many advantages over other clustering algorithms. First, it does not require a fixed number of clusters at all. It also identifies outliers as noise, unlike mean shift, which simply groups data points into clusters even if they are very different. Additionally, it is capable of finding clusters of any size and shape very well.
The main disadvantage of DBSCAN is that it does not perform as well as other clustering algorithms when the density of clusters is different. This is because the settings of the distance threshold ε and minPoints used to identify neighborhood points will change with clusters when density changes. This drawback also arises in very high-dimensional data, as the distance threshold ε again becomes difficult to estimate.
A major drawback of K-Means is its simple use of cluster center means. From the diagram below we can see why this is not the best approach. On the left, you can see very clearly that there are two circular clusters with different radii, centered on the same mean. K-Means cannot handle this situation because the means of these clusters are very close. K-Means also fails in cases where the clusters are not circular, again due to using the mean as the cluster center.
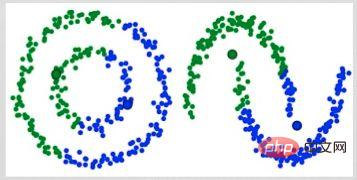
Two Failure Cases of K-Means
Gaussian Mixture Models (GMMs) give us more flexibility than K-Means. For GMMs, we assume that the data points are Gaussian distributed; this is a less restrictive assumption than using the mean to assume that they are circular. This way, we have two parameters to describe the shape of the cluster: mean and standard deviation! Taking 2D as an example, this means that the clusters can take any kind of elliptical shape (since we have standard deviations in both x and y directions). Therefore, each Gaussian distribution is assigned to a single cluster.
To find the Gaussian parameters (such as mean and standard deviation) of each cluster, we will use an optimization algorithm called Expectation Maximum (EM). Look at the diagram below, which is an example of a Gaussian fit to a cluster. We can then continue the process of maximum expectation clustering using GMMs.
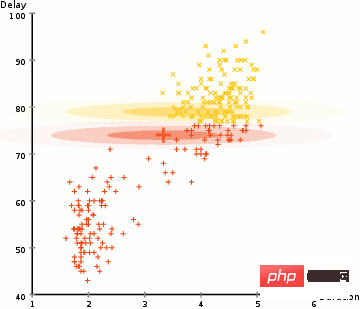
There are two key advantages to using GMMs. First, GMMs are more flexible than K-Means in terms of cluster covariance; because of the standard deviation parameter, clusters can take on any elliptical shape instead of being restricted to circles. K-Means is actually a special case of GMM, where the covariance of each cluster is close to 0 in all dimensions. Second, because GMMs use probabilities, there can be many clusters per data point. So if a data point is in the middle of two overlapping clusters, we can define its class simply by saying that X percent of it belongs to class 1 and Y percent to class 2. That is, GMMs support hybrid qualifications.
Hierarchical clustering algorithms are actually divided into two categories: top-down or bottom-up. Bottom-up algorithms first treat each data point as a single cluster and then merge (or aggregate) two clusters successively until all clusters are merged into a single cluster containing all data points. Therefore, bottom-up hierarchical clustering is called agglomerative hierarchical clustering or HAC. This hierarchy of clusters is represented by a tree (or dendrogram). The root of the tree is the only cluster that collects all samples, and the leaves are the clusters with only one sample. Before going into the algorithm steps, please see the legend below.
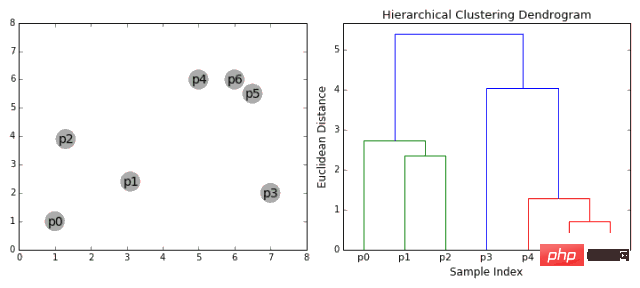
Agglomerative hierarchical clustering
Hierarchical clustering does not require us to specify the number of clusters, we can even choose which number of clusters looks best since we are building a tree. Additionally, the algorithm is not sensitive to the choice of distance metric; they all perform equally well, whereas for other clustering algorithms the choice of distance metric is crucial. A particularly good example of hierarchical clustering methods is when the underlying data has a hierarchical structure and you want to restore the hierarchy; other clustering algorithms cannot do this. Unlike the linear complexity of K-Means and GMM, these advantages of hierarchical clustering come at the cost of lower efficiency since it has a time complexity of O(n³).
When our data can be represented as a network or graph (graph), we can use the graph community detection method to complete clustering. In this algorithm, a graph community is usually defined as a subset of vertices that are more closely connected than other parts of the network.
Perhaps the most intuitive case is social networks. The vertices represent people, and the edges connecting the vertices represent users who are friends or fans. However, to model a system as a network, we must find a way to efficiently connect the various components. Some innovative applications of graph theory for clustering include feature extraction of image data, analysis of gene regulatory networks, etc.
Below is a simple diagram showing 8 recently viewed websites, connected based on links from their Wikipedia pages.
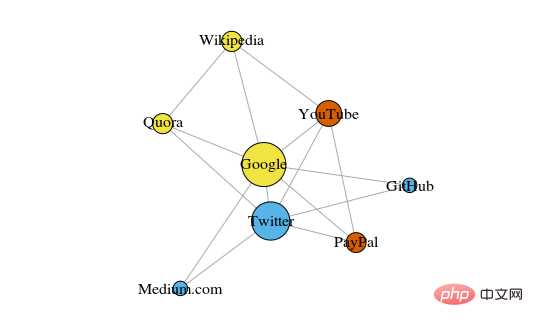
The color of these vertices indicates their group relationship, and the size is determined based on their centrality. These clusters also make sense in real life, where the yellow vertices are usually reference/search sites and the blue vertices are all online publishing sites (articles, tweets, or code).
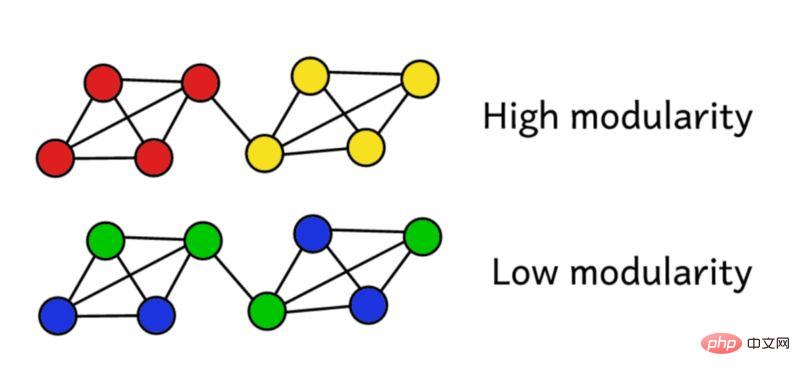
Suppose we have clustered the network into groups. We can then use this modularity score to evaluate the quality of the clustering. A higher score means we segmented the network into "accurate" groups, while a low score means our clustering is closer to random. As shown in the figure below:
Modularity can be calculated using the following formula:
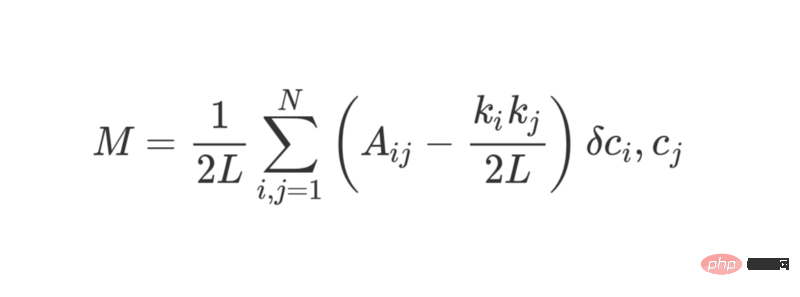
where L represents the edge in the network The quantities, k_i and k_j refer to the degree of each vertex, which can be found by adding up the terms of each row and column. Multiplying the two and dividing by 2L represents the expected number of edges between vertices i and j when the network is randomly assigned.
Overall, the terms in parentheses represent the difference between the true structure of the network and the expected structure when combined randomly. Studying its value shows that it returns the highest value when A_ij = 1 and ( k_i k_j ) / 2L is small. This means that when there is an "unexpected" edge between fixed points i and j, the resulting value is higher.
The last δc_i, c_j is the famous Kronecker δ function (Kronecker-delta function). Here is its Python explanation:

The modularity of the graph can be calculated through the above formula, and the higher the modularity, the better the degree to which the network is clustered into different groups. Therefore, the best way to cluster the network can be found by looking for maximum modularity through optimization methods.
Combinatorics tells us that for a network with only 8 vertices, there are 4140 different clustering methods. A network of 16 vertices would be clustered in over 10 billion ways. The possible clustering methods of a network with 32 vertices will exceed 128 septillion (10^21); if your network has 80 vertices, the number of possible clustering methods has exceeded the number of clustering methods in the observable universe. number of atoms.
We must therefore resort to a heuristic that works well at evaluating the clusters that yield the highest modularity scores, without trying every possibility. This is an algorithm called Fast-Greedy Modularity-Maximization, which is somewhat similar to the agglomerative hierarchical clustering algorithm described above. It’s just that Mod-Max does not fuse groups based on distance, but fuses groups based on changes in modularity.
Here's how it works:
Community detection is a popular research field in graph theory. Its limitations are mainly reflected in the fact that it ignores some small clusters and is only applicable to structured graph models. But this type of algorithm has very good performance in typical structured data and real network data.
The above are the 6 major clustering algorithms that data scientists should know! We’ll end this article by showing visualizations of various algorithms!
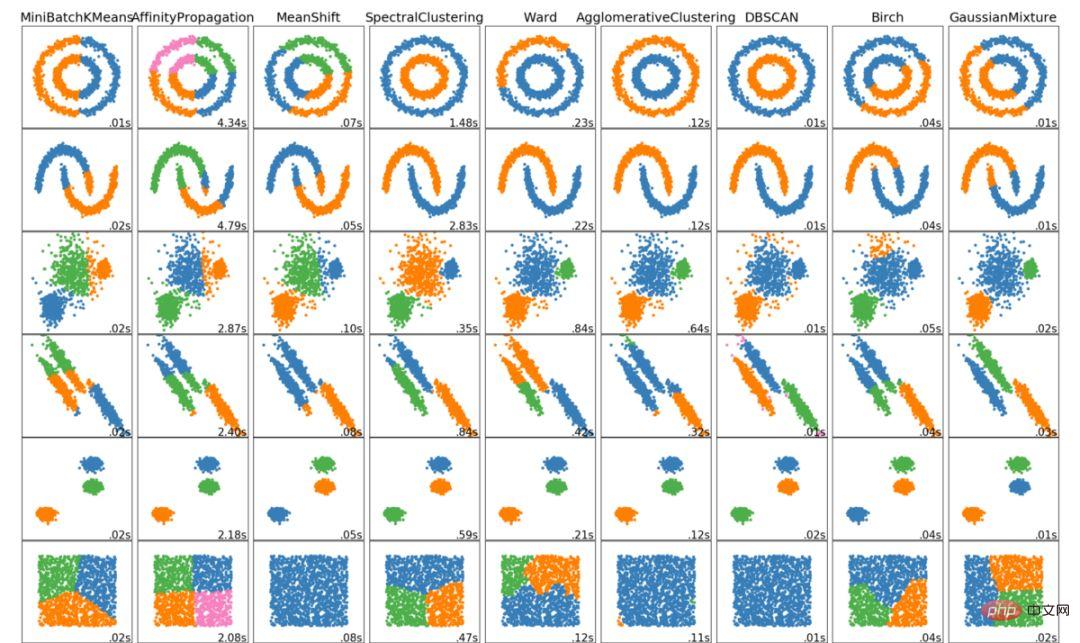
The above is the detailed content of Six clustering algorithms that data scientists must know. For more information, please follow other related articles on the PHP Chinese website!
 Page replacement algorithm
Page replacement algorithm
 Comparative analysis of vscode and visual studio
Comparative analysis of vscode and visual studio
 The difference between Javac and Java
The difference between Javac and Java
 Win10 does not support the disk layout solution of Uefi firmware
Win10 does not support the disk layout solution of Uefi firmware
 Detailed explanation of imp command in oracle
Detailed explanation of imp command in oracle
 The difference between counta and count
The difference between counta and count
 What is a Bitcoin Futures ETF?
What is a Bitcoin Futures ETF?
 How to solve error1
How to solve error1




