
Editor | Radish Skin
Prediction of enzyme kinetic parameters is crucial for the design and optimization of enzymes in biotechnology and industrial applications. However, current prediction tools are Limited performance on various tasks limits their practical applications.
Researchers from the Chinese Academy of Sciences recently proposed UniKP, a unified framework based on pre-trained language models that can be used to predict enzyme kinetic parameters, including enzyme turnover number (kcat), Michaelis-Menten constant (Km) and catalytic efficiency (kcat/Km), these parameters were obtained from the protein sequence and substrate structure.
A two-layer framework based on UniKP (EF-UniKP) is also proposed, which is capable of stably predicting kcat values taking into account environmental factors such as pH and temperature. At the same time, the research team also systematically explored four representative reweighting methods, successfully reducing prediction errors in high-value prediction tasks.
The study is titled "UniKP: a unified framework for the prediction of enzyme kinetic parameters" and was published in the journal "Nature Communications" on December 11, 2023.

Studying the catalytic efficiency of enzymes towards specific substrates is an important issue in biology and has a profound impact on enzyme evolution, metabolic engineering and synthetic biology. In vitro experimental data measuring kcat and Km, as well as the maximum turnover rate and Michaelis-Menten constant, can be used as indicators to measure the efficiency of enzymes in catalyzing specific reactions and to compare the relative catalytic activities of different enzymes.
At present, the measurement of enzyme kinetic parameters mainly relies on experimental measurement, which is time-consuming, costly and labor-intensive, resulting in a small database of experimentally measured kinetic parameter values. For example, the sequence database UniProt contains over 230 million enzyme sequences, while the enzyme databases BRENDA and SABIO-RK contain tens of thousands of experimentally measured kcat values. The integration of Uniprot identifiers in these enzyme databases facilitates the connection between measured parameters and protein sequences. However, the scale of these connections is still much smaller compared to the number of enzyme sequences, limiting progress in downstream applications such as directed evolution and metabolic engineering.
Enzyme kinetic parameter prediction framework
In this study, researchers from the Chinese Academy of Sciences proposed a new framework called UniKP, which is based on pre-training Language model designed to improve the accuracy of predicting enzyme kinetic parameters. These parameters include kcat, Km and kcat/Km, which can be predicted given the enzyme sequence and substrate structure. The researchers conducted a comprehensive comparison of 16 different machine learning models and 2 deep learning models and found that UniKP performed well in terms of prediction accuracy. This research is expected to provide new tools and methods for research and applications in the field of enzyme kinetics.
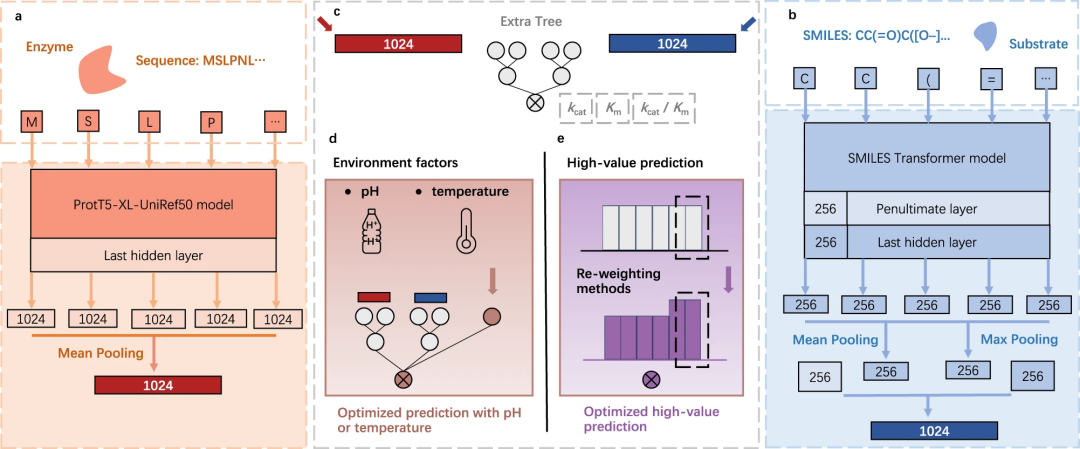
Illustration: UniKP overview. (Source: paper)
Compared with the previous state-of-the-art model DLKcat, UniKP shows superior performance in the kcat prediction task, with an average coefficient of determination of 0.68, an improvement of 20%. The researchers speculate that the pretrained models contributed significantly to UniKP's performance by using unsupervised information from the entire database to create easy-to-learn representations of enzyme sequences and substrate structures.
Analysis of model learning showed that protein information has a dominant role, possibly due to the complexity of the enzyme structure compared to the substrate structure. Furthermore, UniKP can effectively capture small differences in kcat values between enzymes and their mutants, including experimentally measured cases, which is crucial for enzyme design and modification. The difference between the R^2 of UniKP predictions and the R^2 of the gmean method for high- and low-identity regions demonstrates UniKP's ability to extract deeper interconnected information and thus perform well in these tasks. Higher forecast accuracy.
Two-layer framework EF-UniKP
Most current models do not consider environmental factors, which is a key limitation in simulating real experimental conditions. To solve this problem, the researchers proposed a two-layer framework EF-UniKP, which takes environmental factors into account. Based on two newly constructed datasets with pH and temperature information respectively, EF-UniKP shows improved performance compared to the initial UniKP. This is an accurate, high-throughput, organism-independent and context-dependent kcat prediction. Additionally, this approach has the potential to be expanded to include other factors such as co-substrate and NaCl concentration.
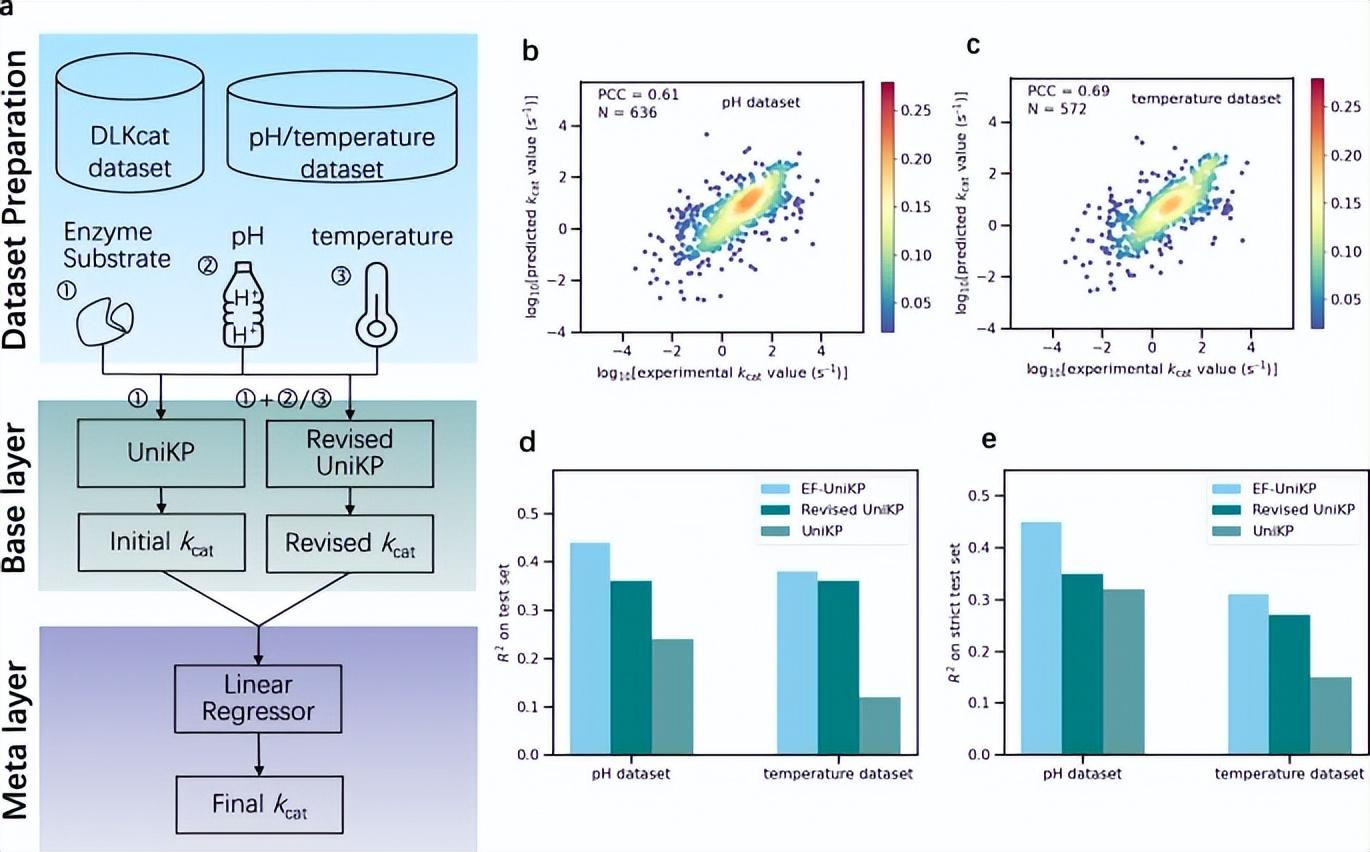
Illustration: Two-layer framework considering environmental factors. (Source: paper)
However, existing models do not consider the interaction between these factors due to a lack of comprehensive data. As experimental techniques advance, including biocast laboratory automation and continuous evolution methods, researchers anticipate a proliferation of enzyme kinetic data. This influx has not only enriched the field but also improved the accuracy of predictive models.
Due to the high imbalance of the kcat dataset, resulting in higher errors in high kcat value predictions, the team systematically explored four representative reweighting methods to alleviate this problem. The results show that the hyperparameter settings of each method are critical to improving high kcat value predictions.
The team confirmed the strong generality of the current framework in terms of Michaelis constant (Km) prediction and kcat/Km prediction. UniKP achieves state-of-the-art performance in predicting Km values and, more impressively, outperforms the combined results of current state-of-the-art models in predicting kcat/Km values. Furthermore, the researchers validated the UniKP framework based on experimentally measured kcat/Km values and kcat/Km values calculated using kcat and Km prediction models on the kcat/Km dataset.
It is worth noting that the correlation observed between the values derived from UniKP kcat / UniKP Km and the experimental kcat / Km is relatively low (PCC = −0.01). This difference may be due to the different data sets used in building the respective models, thus requiring the development of a different model to predict kcat/Km values. In the future, with the emergence of unified data sets containing kcat and Km values, it is expected that the computational output of the kcat and Km models will be closely consistent with the output generated by the kcat/Km dedicated model.
Specific application in enzyme mining and evolution
The application of UniKP in the mining and directed evolution of tyrosine ammonia lyase (TAL) enzyme has demonstrated its ability to revolutionize synthetic biology and potential for biochemical research. This study shows that UniKP effectively recognizes highly active TALs and rapidly improves the catalytic efficiency of existing TALs, with RgTAL-489T having a kcat/Km value 3.5 times higher than that of the wild-type enzyme.
Furthermore, the derived framework EF-UniKP was always able to identify highly active TAL enzymes with extremely high accuracy, with kcat/Km values 2.6-fold higher for TrTAL from Tephrocybe rancida than the wild-type enzyme. The results showed that the kcat and kcat/Km values of the five sequences exceeded those of the wild-type enzyme.
By accelerating the enzyme discovery and optimization process, UniKP is expected to become a powerful tool for advancing biocatalysis, drug discovery, metabolic engineering and other fields that rely on enzyme-catalyzed processes.
Limitations and Outlook
However, the current version of UniKP still has some limitations. For example, while UniKP is able to differentiate between experimentally measured kcat values of an enzyme and its variants, the predicted kcat values are not accurate enough. This may be due to insufficient data sets compared to the number of known protein sequences and substrate structures.
While the reweighting method can alleviate the prediction bias caused by the unbalanced kcat dataset to a certain extent (about 6.5% improvement), more can be achieved through synthetic minority oversampling techniques and other sample synthesis methods. Significant improvement.
A central goal of synthetic biology is the development of digital cells that will revolutionize the way scientists study biology. A key prerequisite for this study is the careful determination of enzymatic parameters for all enzymes within the pathway. Artificial intelligence-assisted tools shed light on this challenge, providing a high-throughput method for predicting enzyme kinetics.
Although the UniKP predictor error is reduced compared to earlier models, inaccuracy remains a significant obstacle to building accurate metabolic models. Incorporating an increasing number of experimentally determined kcat and Km values can improve model accuracy.
Next, the researchers plan to combine state-of-the-art algorithms such as transfer learning, reinforcement learning, and other small-shot learning algorithms to effectively handle imbalanced data sets. And, the team aims to explore additional applications, including enzyme evolution and global analysis of organisms.
Paper link: https://www.nature.com/articles/s41467-023-44113-1
The above is the detailed content of Chinese Academy of Sciences team creates a unified framework for improving prediction accuracy of enzyme kinetic parameters. For more information, please follow other related articles on the PHP Chinese website!




