
The properties of a material are determined by the arrangement of its atoms. However, existing methods for obtaining such arrangements are either too expensive or ineffective for many elements.
Now, researchers in UC San Diego’s Department of Nanoengineering have developed an artificial intelligence algorithm that can predict the structure and dynamics of any material, whether existing or new, almost instantly characteristic. The algorithm, known as M3GNet, was used to develop the matterverse.ai database, which contains more than 31 million yet-to-be-synthesized materials whose properties are predicted by machine learning algorithms. Matterverse.ai facilitates the discovery of new technological materials with exceptional properties.
The research is titled "A universal graph deep learning interatomic potential for the periodic table" and was published in "Nature Computational Science on November 28, 2022 "superior.
Paper link: https://www.nature.com/articles/s43588-022-00349-3
For large-scale materials research, efficient, linearly scaled interatomic potentials (IAPs) are needed to describe potential energy surfaces (PES) in terms of many-body interactions between atoms. However, most IAPs today are customized for a narrow range of chemicals: usually a single element or no more than four or five elements at most.
Recently, machine learning of PES has emerged as a particularly promising approach to IAP development. However, no studies have demonstrated a universally applicable IAP across the periodic table and across all types of crystals.
Over the past decade, the emergence of efficient and reliable electronic structure codes and high-throughput automation frameworks has led to the development of large federated databases of computational materials data. A large amount of PES data has been accumulated during structural relaxation, i.e., intermediate structures and their corresponding energies, forces, and stresses, but less attention has been paid to these data.
"Similar to proteins, we need to understand the structure of a material to predict its properties." said Shyue Ping Ong, the study's lead author. "What we need is AlphaFold for materials."
AlphaFold is an artificial intelligence algorithm developed by Google DeepMind to predict protein structures. To build the material equivalent, Ong and his team combined graph neural networks with many-body interactions to build a deep learning architecture that is universal across all elements of the periodic table. , work with high precision.
Mathematical graphs are natural representations of crystals and molecules, with nodes and edges representing atoms and the bonds between them respectively. Traditional material graph neural network models have proven to be very effective for general material property prediction, but lack physical constraints and are therefore unsuitable for use as IAPs. The researchers developed a material graph architecture that explicitly incorporates multibody interactions. Model development is inspired by traditional IAP, and in this work, the focus will be on the integration of three-body interactions (M3GNet).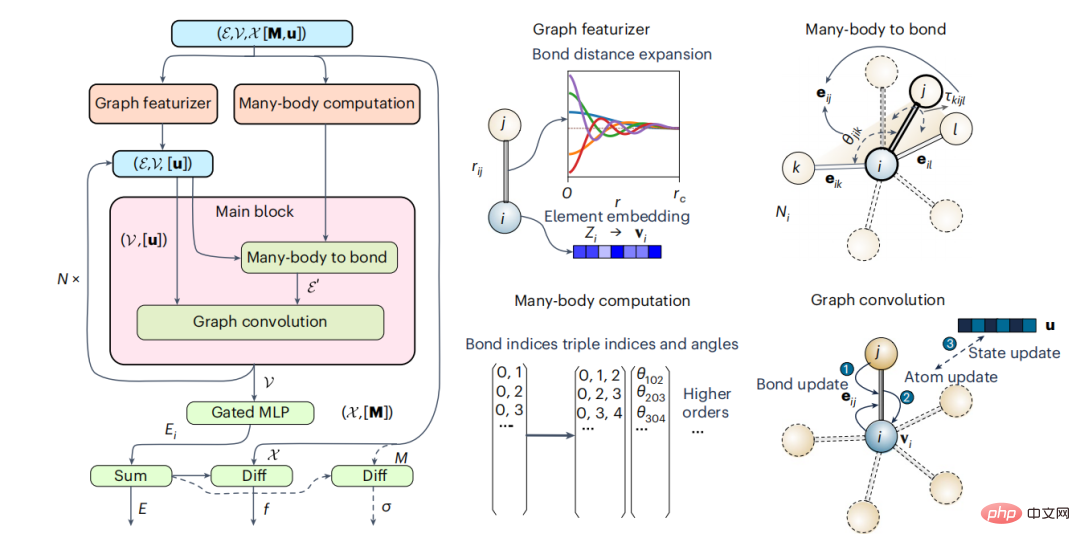
Figure 1: Schematic diagram of multi-body graph potential and main calculation blocks. (Source: Paper)
Benchmarking on IAP Dataset
As an initial benchmark, the researchers chose Ong and colleagues previously generated diverse DFT data sets of elemental energies and forces for face-centered cubic (fcc) nickel, fcc copper, body-centered cubic (bcc) lithium, bcc molybdenum, diamond silicon, and diamond germanium.Table 1: Error comparison between M3GNet model and existing models EAM, MEAM, NNP and MTP on single-element data sets. (Source: paper)
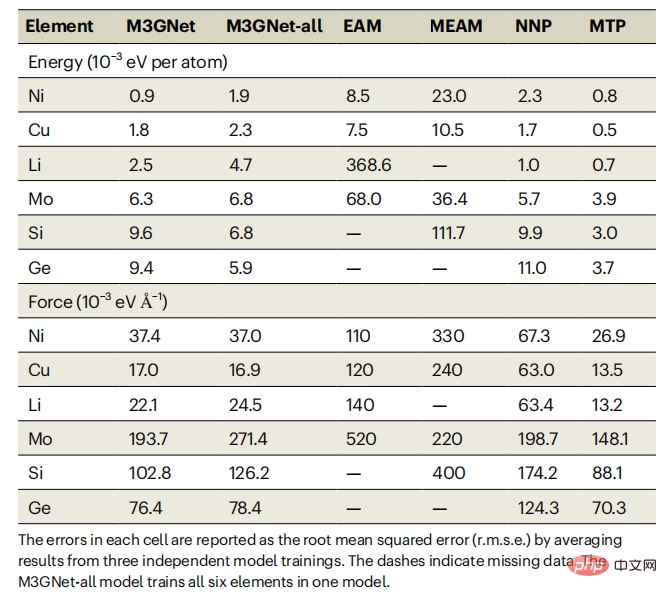
As can be seen from Table 1, M3GNet IAP is significantly better than the classic multi-body potential; their performance is also comparable to ML-IAP based on the local environment. It should be noted that although ML-IAP can achieve slightly smaller energy and force errors than M3GNet IAP, its flexibility in handling multi-element chemistry will be greatly reduced, since adding multiple elements to ML-IAP often results in Number of combined explosion regression coefficients and corresponding data requirements. In contrast, the M3GNet architecture represents the elemental information of each atom (node) as a learnable embedding vector. Such a framework is easily extended to multicomponent chemistry.
Like other GNNs, the M3GNet framework is able to capture long-range interactions without increasing the cutoff radius for bond construction. At the same time, unlike previous GNN models, the M3GNet architecture still maintains continuous changes in energy, force, and stress as the number of bonds changes, which is a key requirement for IAP.
Universal IAP for the periodic table
To develop an IAP for the entire periodic table, the team used the world’s largest DFT crystal structure One of the open databases on relaxation (Materials Project).
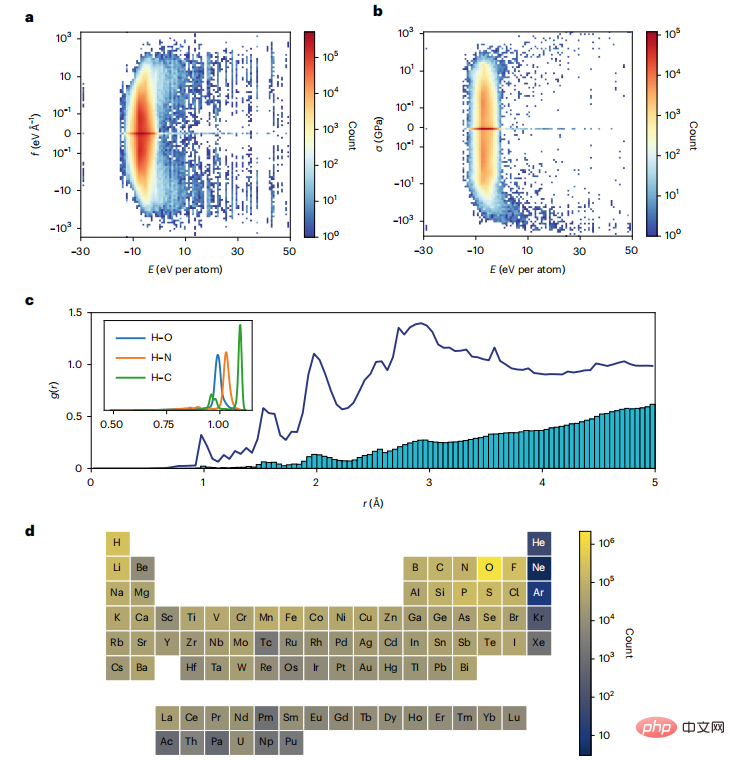
Figure 2: Distribution of MPF.2021.2.8 data set. (Source: paper)
In principle, IAP can train energy only, or a combination of energy and force. In practice, M3GNet IAP (M3GNet-E) trained only on energy cannot achieve reasonable accuracy in predicting force or stress, with mean absolute error (MAE) even larger than the mean absolute deviation of the data. The M3GNet models trained with energy force (M3GNet-EF) and energy force stress (M3GNet-EFS) obtained relatively similar energy and force MAE, but the stress MAE of M3GNet-EFS was about half that of the M3GNet-EF model.
For applications involving lattice changes, such as structural relaxation or NpT molecular dynamics simulations, accurate stress prediction is necessary. The findings indicate that including all three properties (energy, force, and pressure) in model training is crucial to obtain practical IAP. The final M3GNet-EFS IAP (hereafter referred to as the M3GNet model) achieved an average of 0.035eV per atom, with average energy, force, and pressure test MAEs of 0.072eVÅ−1 and 0.41GPa, respectively.
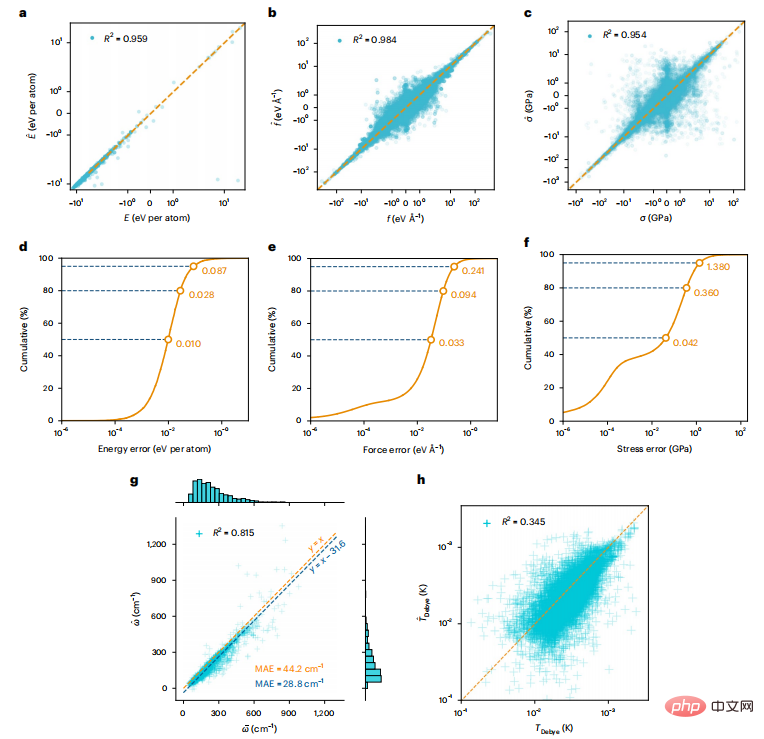
Figure 3: Model predictions on the test dataset compared to DFT calculations.
On the test data, the model predictions and DFT ground truth match well, as revealed by the high linearity and R2 value of the linear fit between DFT and model predictions. The cumulative distribution of model errors shows that 50% of the data have energy, force, and stress errors less than 0.01 eV, 0.033 eVÅ−1, and 0.042 GPa per atom, respectively. The Debye temperature calculated by M3GNet is less accurate, which can be attributed to M3GNet's relatively poor prediction of shear modulus; however, the bulk modulus prediction is reasonable.
The M3GNet IAP is then applied to simulate material discovery workflows where the final DFT structure is unknown a priori. M3GNet relaxation was performed on initial structures from a test dataset of 3,140 materials. Energy calculations of the M3GNet relaxed structures yielded a MAE of 0.035 eV per atom, and 80% of the materials had an error less than 0.028 eV per atom. The error distribution of the relaxed structure using M3GNet is close to that of the known DFT final structure, indicating the potential of M3GNet to accurately help obtain the correct structure. Generally speaking, the relaxation of M3GNet converges quickly.
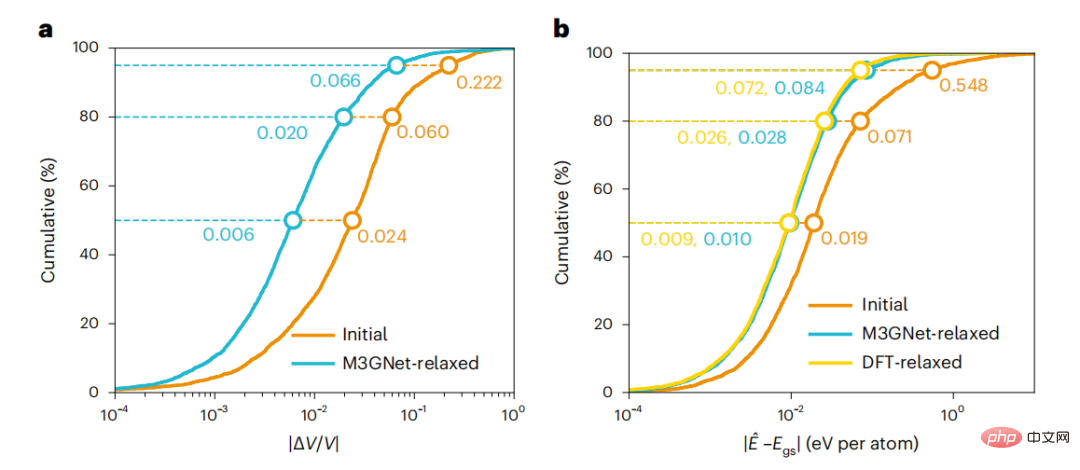
Figure 4: Relaxed crystal structure using M3GNet. (Source: paper)
New material discovery
M3GNet can accurately and quickly relax any crystal structures and predict their energies, making them ideal for large-scale materials discovery. The researchers generated 31,664,858 candidate structures as starting points, using M3GNet IAP to relax the structures and calculate the signed energy distance to the Materials Project convex hull (Ehull-m); 1,849,096 materials had Ehull-m less than 0.01 eV per atom.
As a further evaluation of M3GNet’s performance in materials discovery, the researchers calculated the discovery rate as uniformly sampling 1000 structures of DFT-stable materials from approximately 1.8 million Ehull-m materials smaller than 0.001 eV/atom (Ehull− dft ≤ 0). The discovery rate remains close to 1.0 up to the Ehull-m threshold of approximately 0.5 eV per atom, and remains at a reasonably high value of 0.31 at the most stringent threshold of 0.001 eV per atom.
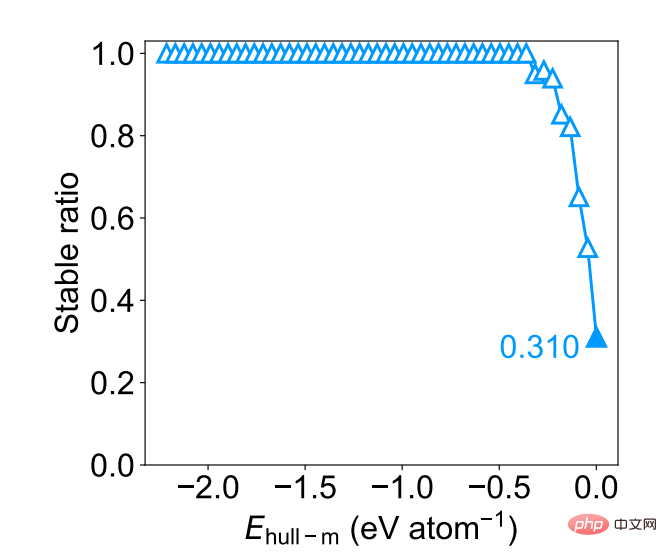
Figure 5: DFT stability ratio as a function of Ehull−m threshold for a uniform sample of 1000 structures. (Source: paper)
For this material set, the researchers also compared the DFT relaxation time cost with and without M3GNet pre-relaxation. The results show that without M3GNet pre-relaxation, the DFT relaxation time cost is approximately 3 times that of M3GNet pre-relaxation.
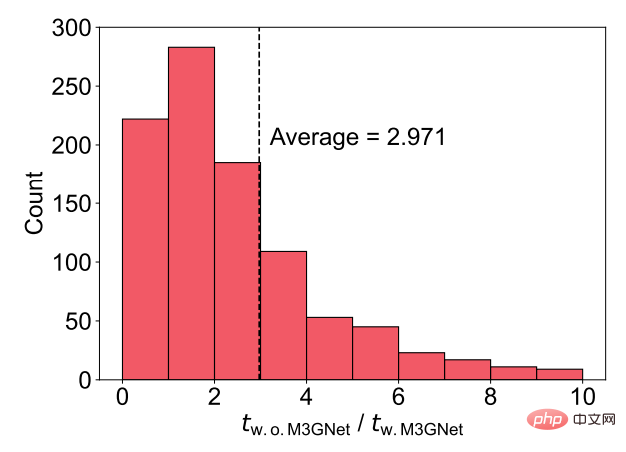
Figure 6: DFT acceleration using M3GNet pre-relaxation. (Source: paper)
Of the 31 million materials in matterverse.ai today, more than 1 million are expected to be potentially stable. Ong and his team intend to significantly expand not only the number of materials, but also the number of attributes ML can predict, including high-value attributes for small data volumes using the multi-fidelity approach they previously developed.
In addition to structural relaxation, M3GNet IAP also has extensive applications in material dynamic simulation and performance prediction.
"For example, we are often interested in the rate of diffusion of lithium ions in the electrode or electrolyte of a lithium-ion battery. The faster the diffusion, the faster the battery can be charged or discharged," Ong said. "We have demonstrated that M3GNet IAP can be used to predict the lithium conductivity of materials with high accuracy. We firmly believe that the M3GNet architecture is a transformative tool that can greatly expand our ability to explore new materials chemistry and structure."
To promote the use of M3GNet, the team has released the framework as open source Python code on Github. There are plans to integrate M3GNet IAP as a tool into commercial materials simulation packages.
The above is the detailed content of Common to the entire periodic table of elements, AI instantly predicts material structure and properties. For more information, please follow other related articles on the PHP Chinese website!
 Recommended computer hardware testing software rankings
Recommended computer hardware testing software rankings
 Introduction to software development tools
Introduction to software development tools
 How many people can you raise on Douyin?
How many people can you raise on Douyin?
 linux switch user command
linux switch user command
 The difference between JD.com's self-operated flagship store and its official flagship store
The difference between JD.com's self-operated flagship store and its official flagship store
 What are the common testing techniques?
What are the common testing techniques?
 How to take screenshots on Huawei mate60pro
How to take screenshots on Huawei mate60pro
 The difference between arrow functions and ordinary functions
The difference between arrow functions and ordinary functions




