
The first step is to install geatpy, just use the pip3 command to install it:
pip3 install geatpy
The following prompt appears: Successful installation:
Most of the data in geatpy is stored and calculated using numpy arrays , below I will introduce how the concepts in genetic algorithms are represented by numpy data, and the meaning of rows and columns.
The most important thing in the genetic algorithm is the chromosome representation of the individual. In geatpy, the population chromosome is represented by Chrom. This is a two-dimensional array, in which each row corresponds to an individual. Chromosome encoding, the structure of Chrom is as follows: lind represents the length of the encoding, and Nind represents the size of the population (number of individuals).

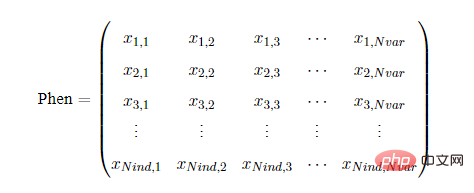
The population phenotype refers to the gene phenotype matrix Phen obtained after decoding the population chromosome matrix Chrom. Each row corresponds to a Individual, each column corresponds to a decision variable. The structure of Phen is as follows: Nvar represents the number of variables
The value of Phen is related to the decoding method used. Geatpy provides a decoding method for converting binary/Gray code encoding to decimal integer or real number. In addition, Geatpy can also use a "real-valued encoding" population that does not require decoding. Each bit of the chromosome of this population corresponds to the actual value of the decision variable, that is, Phen is equivalent to Chrom under this encoding method.
Geatpy uses the numpy array type matrix to store the objective function value of the population. Generally named ObjV, each row corresponds to each individual, so it has the same number of rows as Chrom; each column corresponds to an objective function, so for a single objective function, ObjV will only have 1 column; and for multiple objective functions, ObjV will have Multi-column, the representation of ObjV is as follows:
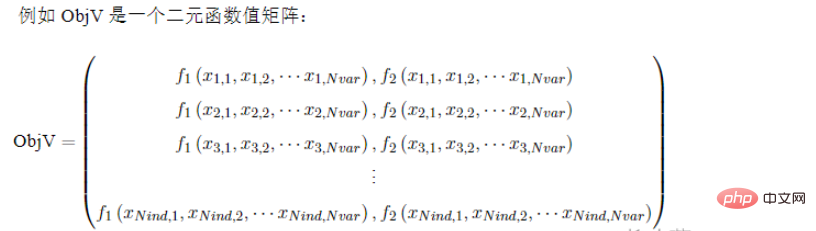
Note: Fitness in Geatpy follows "minimum fitness is 0 "The agreement. 2.5 Violation of Constraint Degree Matrix
 If an element of the CV matrix is less than or equal to 0, it means that the element corresponds to The individual satisfies the corresponding constraints. If it is greater than 0, it means that the constraint condition is violated. If the value is greater than 0, the greater the value, the higher the degree of violation of the constraint by the individual. Geatpy provides two methods for handling constraints, one is the penalty function method and the other is the feasibility rule. When using feasibility rules to deal with constraints, you need to use the CV matrix.
If an element of the CV matrix is less than or equal to 0, it means that the element corresponds to The individual satisfies the corresponding constraints. If it is greater than 0, it means that the constraint condition is violated. If the value is greater than 0, the greater the value, the higher the degree of violation of the constraint by the individual. Geatpy provides two methods for handling constraints, one is the penalty function method and the other is the feasibility rule. When using feasibility rules to deal with constraints, you need to use the CV matrix.
2.6 Decoding Matrix
When only using the library functions of the toolbox without using the object-oriented evolutionary algorithm framework provided by Geatpy, the decoding matrix can be used alone. If the object-oriented evolutionary algorithm framework provided by Geatpy is used, the decoding matrix can be used in conjunction with a string Encoding that stores the population chromosome encoding method.
Currently there are three Encodings in Geatpy, which are:
注:’RI’和’P’编码的染色体都不需要解码,染色体上的每一位本身就代表着决策变量的真实值,因此“实整数编码”和“排列编码”可统称为“实值编码”
以BG编码为例,我们展示一下编译矩阵FieldD。FieldD的结构如下:
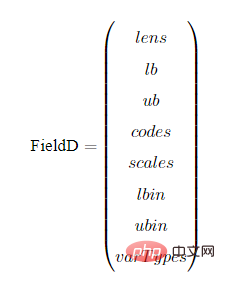
其中,lens,lb,ub,codes,scales,lbin,ubin,varTypes都是行向量,其长度等于决策变量的个数。
lens:代表以条染色体中,每个子染色体的长度。
lb:代表每个变量的上界
ub:代表每个变量的下界
codes:代表染色体字串用的编码方式,[1,0,1]代表第一个变量用的格雷编码,第二个变量用的二进制编码,第3个变量用的格雷编码。
scales:指明每个子串用的是算术刻度还是对数刻度。scales[i] = 0为算术刻度,scales[i] = 1为对数刻度(对数刻度很少用,可以忽略。)
lbin:代表变量上界是否包含其范围边界。0代表不包含,1代表包含。‘[ ’和 ‘(’ 的区别
ubin:代表变量下界是否包含其范围边界。0代表不包含,1代表包含。
varTypes:代表决策变量的类型,元素为0表示对应位置的决策变量是连续型变量;1表示对应的是离散型变量。
例如:有以下一个译码矩阵

它表示待解码的种群染色体矩阵Chrom解码后可以表示成3个决策变量,每个决策变量的取值范围分别是[1,10], [2,9], [3,15]。其中第一第二个变量采用的是二进制编码,第三个变量采用的是格雷编码,且第一、第三个决策变量为连续型变量;第二个为离散型变量。
#通过种群染色体chrom和译码矩阵FieldD,可解码成种群表现型矩阵。 import geatpy as ea Phen = ea.bs2ri(Chrom, FieldD)
在使用Geatpy进行进化算法编程时,常常建立一个进化追踪器(如pop_trace)来记录种群在进化的过程中各代的最优个体,尤其是采用无精英保留机制时,进化追踪器帮助我们记录种群在进化过程中的最优个体。待进化完成后,再从进化追踪器中挑选出“历史最优”的个体。这种进化记录器有多种,其中一种是numpy的array类型的,结构如下:其中MAXGEN是种群进化的代数(迭代次数)。
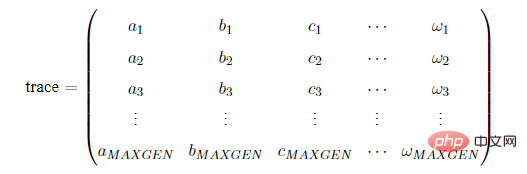
trace的每一列代表不同的指标,比如第一列记录各代种群的最佳目标函数值,第二列记录各代种群的平均目标函数值…trace的每一行对应每一代,如第一行代表第一代,第二行代表第二代…另外一种进化记录器是一个列表,列表中的每一个元素都是一个拥有相同数据类型的数据。比如在Geatpy的面向对象进化算法框架中的pop_trace,它是一个列表,列表中的每一个元素都是历代的种群对象。
在Geatpy提供的面向对象进化算法框架中,种群类(Population)是一个存储着与种群个体相关信息的类。它有以下基本属性:
sizes : int -种群规模,即种群的个体数目。
ChromNum : int -染色体的数目,即每个个体有多少条染色体。
Encoding : str -染色体编码方式。
Field : array -译码矩阵,可以是FieldD或FieldDR。
Chrom : array -种群染色体矩阵,每一行对应一个个体的一条染色体。
Lind : int -种群染色体长度。
ObjV : array -种群目标函数值矩阵。
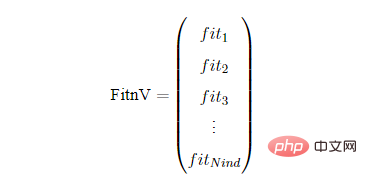
FitnV : array -种群个体适应度列向量。
CV : array -种群个体违反约束条件程度的矩阵。
Phen : array -种群表现型矩阵。
可以直接对种群对象进行提取个体、个体合并等操作,比如pop1和pop2是两个种群对象,则通过语句“pop3 = pop1 + pop2”,即可把两个种群的个体合并,得到一个新的种群。在合并的过程中,实际上是把种群的各个属性进行合并,然后用合并的数据来生成一个新的种群(详见Population.py)。又比如执行语句“pop3 = pop1[[0]]”,可以把种群的第0号个体抽取出来,得到一个新的只有一个个体的种群对象pop3。值得注意的是,种群的这种个体抽取操作要求下标必须为列表或是Numpy array类型的行向量,不能是标量(详见Population.py)
PsyPopulation类是Population的子类,它提供Population类所不支持的多染色体混合编码。它有以下基本属性:
sizes : int -种群规模,即种群的个体数目。
ChromNum : int -染色体的数目,即每个个体有多少条染色体。
Encodings : list -存储各染色体编码方式的列表。
Fields : list -存储各染色体对应的译码矩阵的列表。
Chroms : list -存储种群各染色体矩阵的列表。
Linds : list -存储种群各染色体长度的列表。
ObjV : array -种群目标函数值矩阵。
FitnV : array -种群个体适应度列向量。
CV : array -种群个体违反约束条件程度的矩阵。
Phen : array -种群表现型矩阵。
可见PsyPopulation类基本与Population类一样,不同之处是采用Linds、Encodings、Fields和Chroms分别存储多个Lind、Encoding、Field和Chrom。
PsyPopulation类的对象往往与带“psy”字样的进化算法模板配合使用,以实现多染色体混合编码的进化优化。
遗传算法求解以下函数的最小值:
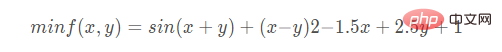
代码实现:
#-*-coding:utf-8-*-
import numpy as np
import geatpy as ea#导入geatpy库
import time
"""============================目标函数============================"""
def aim(Phen):#传入种群染色体矩阵解码后的基因表现型矩阵
x1 = Phen[:, [0]]#取出第一列,得到所有个体的第一个自变量
x2 = Phen[:, [1]]#取出第二列,得到所有个体的第二个自变量
return np.sin(x1 + x2) + (x1 - x2) ** 2 - 1.5 * x1 + 2.5 * x2+1
"""============================变量设置============================"""
x1 = [-1.5, 4]#第一个决策变量范围
x2 = [-3, 4]#第二个决策变量范围
b1 = [1, 1]#第一个决策变量边界,1表示包含范围的边界,0表示不包含
b2 = [1, 1]#第二个决策变量边界,1表示包含范围的边界,0表示不包含
#生成自变量的范围矩阵,使得第一行为所有决策变量的下界,第二行为上界
ranges=np.vstack([x1, x2]).T
#生成自变量的边界矩阵
borders=np.vstack([b1, b2]).T
varTypes = np.array([0, 0])#决策变量的类型,0表示连续,1表示离散
"""==========================染色体编码设置========================="""
Encoding ='BG'#'BG'表示采用二进制/格雷编码
codes = [1, 1]#决策变量的编码方式,两个1表示变量均使用格雷编码
precisions =[6, 6]#决策变量的编码精度,表示解码后能表示的决策变量的精度可达到小数点后6位
scales = [0, 0]#0表示采用算术刻度,1表示采用对数刻度#调用函数创建译码矩阵
FieldD =ea.crtfld(Encoding,varTypes,ranges,borders,precisions,codes,scales)
"""=========================遗传算法参数设置========================"""
NIND = 20#种群个体数目
MAXGEN = 100#最大遗传代数
maxormins = np.array([1])#表示目标函数是最小化,元素为-1则表示对应的目标函数是最大化
selectStyle ='sus'#采用随机抽样选择
recStyle ='xovdp'#采用两点交叉
mutStyle ='mutbin'#采用二进制染色体的变异算子
Lind =int(np.sum(FieldD[0, :]))#计算染色体长度
pc= 0.9#交叉概率
pm= 1/Lind#变异概率
obj_trace = np.zeros((MAXGEN, 2))#定义目标函数值记录器
var_trace = np.zeros((MAXGEN, Lind))#染色体记录器,记录历代最优个体的染色体
"""=========================开始遗传算法进化========================"""
start_time = time.time()#开始计时
Chrom = ea.crtpc(Encoding,NIND, FieldD)#生成种群染色体矩阵
variable = ea.bs2ri(Chrom, FieldD)#对初始种群进行解码
ObjV = aim(variable)#计算初始种群个体的目标函数值
best_ind = np.argmin(ObjV)#计算当代最优个体的序号
#开始进化
for gen in range(MAXGEN):
FitnV = ea.ranking(maxormins * ObjV)#根据目标函数大小分配适应度值
SelCh = Chrom[ea.selecting(selectStyle,FitnV,NIND-1),:]#选择
SelCh = ea.recombin(recStyle, SelCh, pc)#重组
SelCh = ea.mutate(mutStyle, Encoding, SelCh, pm)#变异
# #把父代精英个体与子代的染色体进行合并,得到新一代种群
Chrom = np.vstack([Chrom[best_ind, :], SelCh])
Phen = ea.bs2ri(Chrom, FieldD)#对种群进行解码(二进制转十进制)
ObjV = aim(Phen)#求种群个体的目标函数值
#记录
best_ind = np.argmin(ObjV)#计算当代最优个体的序号
obj_trace[gen,0]=np.sum(ObjV)/ObjV.shape[0]#记录当代种群的目标函数均值
obj_trace[gen,1]=ObjV[best_ind]#记录当代种群最优个体目标函数值
var_trace[gen,:]=Chrom[best_ind,:]#记录当代种群最优个体的染色体
# 进化完成
end_time = time.time()#结束计时
ea.trcplot(obj_trace, [['种群个体平均目标函数值','种群最优个体目标函数值']])#绘制图像
"""============================输出结果============================"""
best_gen = np.argmin(obj_trace[:, [1]])
print('最优解的目标函数值:', obj_trace[best_gen, 1])
variable = ea.bs2ri(var_trace[[best_gen], :], FieldD)#解码得到表现型(即对应的决策变量值)
print('最优解的决策变量值为:')
for i in range(variable.shape[1]):
print('x'+str(i)+'=',variable[0, i])
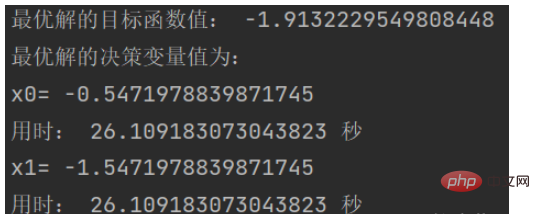
print('用时:', end_time - start_time,'秒')效果图:
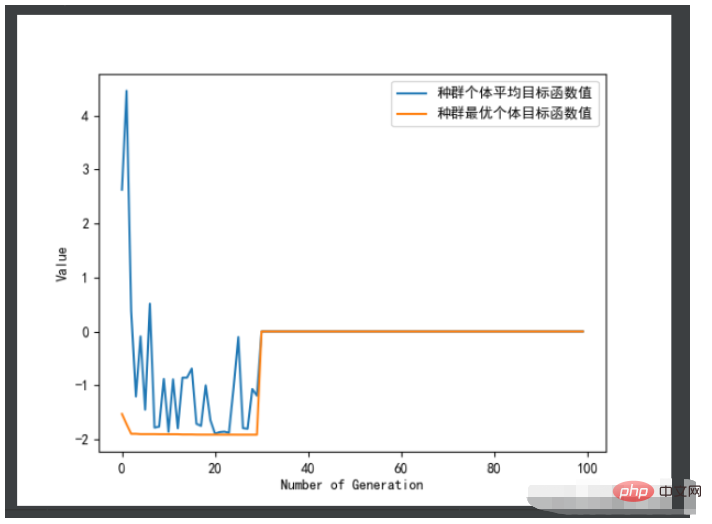
结果如下:
The above is the detailed content of How to install and use python genetic algorithm geatpy. For more information, please follow other related articles on the PHP Chinese website!




