
Translator|Cui Hao
Reviewer|Sun Shujuan
This article explores how TypeDB helps scientists achieve the next breakthrough in medicine, and will pass Instructive code examples and visuals demonstrate the results.

#There is a lot of hype in the biotech world focused on revolutionary drug discovery. After all, the past decade has been a golden age for the field. Compared to the previous decade, 73% more new drugs were approved between 2012 and 2021 - a 25% increase from the previous decade. These include immunotherapies to treat cancer, gene therapies and, of course, the Covid vaccine. It can be seen from these aspects that the pharmaceutical industry is doing well.
But the trend is increasingly worrying. The costs and risks of drug discovery are becoming prohibitive. Up to now, the average cost of bringing a new drug to the market is US$1 billion to US$3 billion, and the average time is 12 to 18 years. At the same time, the average price of a new drug has soared from $2,000 in 2007 to $180,000 in 2021.
That’s why many are pinning their hopes on artificial intelligence (AI), such as statistical machine learning, to help accelerate the development of new drugs, from early target identification to trials. While some compounds have been identified using various machine learning algorithms, these compounds are still in early discovery or preclinical development stages. The promise of artificial intelligence to revolutionize drug discovery remains an exciting but unfulfilled promise.
In order to realize this promise, it is crucial to understand what artificial intelligence really means. In recent years, the term artificial intelligence has become a quite popular term without much technical content. So, what is real artificial intelligence?
Artificial intelligence, as an academic field, has been around since the 1950s, and over time has branched into various types, representing different learning styles. Professor Pedro Domingos describes these types (he calls them "tribes") in his book Masters of Algorithms: connectionists, symbolists, evolutionists, Bayesians and Simulationist.
While Bayesians and connectionists have received much public attention over the past decade, symbolists have not. Semiotics creates realistic representations of the world based on sets of rules for logical reasoning. Symbolic AI systems don’t have the huge publicity that other types of AI enjoy, but they possess unique and important capabilities that other types lack: automated reasoning and knowledge representation.
In fact, the problem of knowledge representation is one of the biggest problems in drug discovery. Existing database software, such as relational or graph databases, struggle to accurately represent and understand the intricacies of biology.
The problem formulated by Drug Discovery is a good example of the need to build unified models for different biomedical data sources (such as Uniprot or Disgenet). At the database level, this means creating data models (some might call these ontologies) that describe myriad complex entities and relationships, such as those between proteins, genes, drugs, diseases, interactions, and more.
This is what TypeDB, an open source database software, aims to achieve - to enable developers to create realistic representations of highly complex domains that computers can use to gain insights.
TypeDB’s type system is based on the concept of entity relationships and represents the data stored in TypeDB. This makes it powerful enough to capture complex biomedical domain knowledge (through type reasoning, nested relations, hyper-relations, rule reasoning, etc.), allowing scientists to gain insights and accelerate drug development time.
This is illustrated by the example of a large pharmaceutical company that struggled for more than five years to model a disease network using Semantic Web standards, but successfully implemented it in just three weeks after migrating to TypeDB achieved this goal.
For example, a biomedical model describing proteins, genes, and diseases written in TypeQL (TypeDB’s query language) looks like this:
define protein sub entity, owns uniprot-id, plays protein-disease-association:protein, plays encode:encoded-protein; gene sub entity, owns entrez-id, plays gene-disease-association:gene, plays encode:encoding-gene; disease sub entity, owns disease-name, plays gene-disease-association:disease, plays protein-disease-association:disease; encode sub relation, relates encoded-protein, relates encoding-gene; protein-disease-association sub relation, relates protein, relates disease; gene-disease-association sub relation, relates gene, relates disease; uniprot-id sub attribute, value string; entrez-id sub attribute, value string; disease-name sub attribute, value string;
For a complete working example, you can Found an open source biomedical knowledge graph on Github. This is loaded from various well-known biomedical resources such as Uniprot, Disgenet, Reactome and others.
With data stored in TypeDB, you can run queries asking questions such as: Which drugs interact with genes related to the SARS virus?
To answer this question, we can use the following query in TypeQL.
match $virus isa virus, has virus-name "SARS"; $gene isa gene; $drug isa drug; ($virus, $gene) isa gene-virus-association; ($gene, $drug) isa drug-gene-interaction;
Running this will cause TypeDB to return data that matches the query conditions. and can be visualized in TypeDB Studio as shown below, which will help understand which related drugs may deserve further investigation.
通过自动推理,TypeDB也可以推断出数据库中不存在的知识。这是通过编写规则来完成的,这些规则构成了TypeDB中模式的一部分。例如,一个规则可以推断出一个基因和一种疾病之间的关联,如果该基因编码的蛋白质与该疾病有关。这样的规则将被写成:
rule inference-example:
when {
(encoding-gene: $gene, encoded-protein: $protein) isa encode;
(protein: $protein, disease: $disease) isa protein-disease-association;
} then {
(gene: $gene, disease: $disease) isa gene-disease-association;
};然后,如果我们要插入以下数据:
TypeDB将能够推断出基因和疾病之间的联系,即使没有插入到数据库中。在这种情况下,以下关系基因-疾病-关联将被推断出来。
match $gene isa gene, has gene-id "2"; $disease isa disease, has disease-name $dn; ; (gene: $gene, disease:$disease) isa gene-disease-assocation;
有了TypeDB对生物医学数据(符号)进行表示,再加上机器学习的上下文知识就可以让整个系统变得更加强大,从而增强洞察力。例如,可以通过药物探索管道发现有希望的目标。
寻找有希望的目标的方法是使用链接预测算法。TypeDB的规则引擎允许这样的ML模型执行,该模型通过推理推断对事实进行学习。这意味着从对平面的、无背景的数据学习转向对推理的、有背景的知识学习。其中一个好处是,根据领域的逻辑规则,预测可以被概括到训练数据的范围之外,并减少所需的训练数据量。
这样一个药物发现的工作流程如下:
1. 查询TypeDB,创建上下文知识的子图,利用TypeDB的全部表达能力。
2. 将子图转化为嵌入(embedding),并将这些嵌入到图学习算法中。
3. 预测结果(例如,作为基因-疾病关联之间的概率分数)可以被插入TypeDB,并用于验证/优先考虑某些目标。
有了数据库中的这些预测,我们可以提出更高层次的问题,利用这些预测与数据库中更广泛的背景知识。比如说:什么是最有可能成为黑色素瘤的基因目标,这些基因编码的蛋白质在黑色素细胞中如何表达?
用TypeQL写,这个问题看起来如下:
match $gene isa gene, has gene-id $gene-id; $protein isa protein; $cell isa cell, has cell-type "melanocytes"; $disease isa disease, has disease-name "melanoma"; ($gene, $protein) isa encode; ($protein, $cell) isa expression; ($gene, $disease) isa gene-disease-association, has prob $p; get $gene-id; sort desc $p;
这个查询的结果将是一个按概率分数排序的基因列表(如图学习者预测的):
{$gid "TOPGENE" isa gene-id;}
{$gid "BESTGENE" isa gene-id;}
{$gid "OTHERTARGET" isa gene-id;}
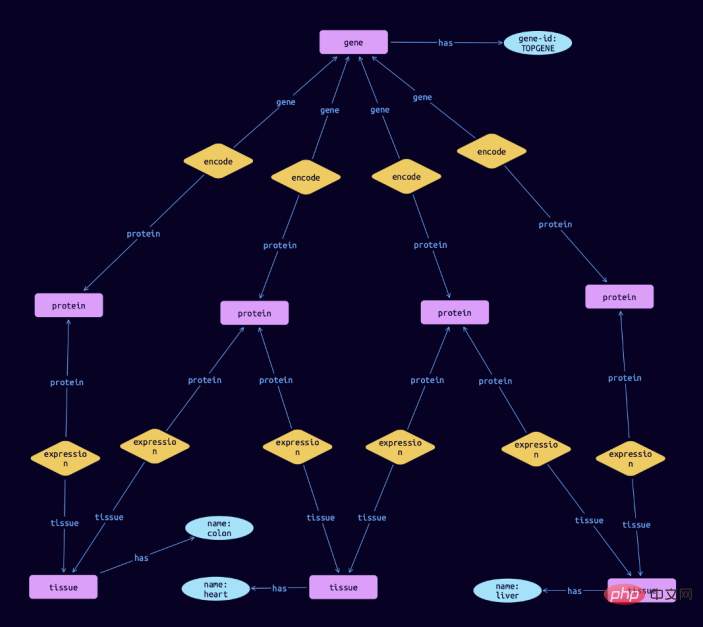
...然后,我们可以进一步研究这些基因,例如通过了解每个基因的生物学背景。比方说,我们想知道TOPGENE基因编码的蛋白质所处的组织。我们可以写下面的查询。
match $gene isa gene, has gene-id $gene-id; $gene-id "TOPGENE"; $protein isa protein; $tissue isa tissue, has name $name; $rel1 ($gene, $protein); $rel2 ($protein, $tissue);
在TypeDB Studio中可视化的结果,可以显示这个基因编码的蛋白质在结肠、心脏和肝脏中的表达:
世界迫切需要创造治疗破坏性疾病的解决方案,希望通过人工智能的创新建立一个更健康的世界,在这个世界中每种疾病都可以被治疗。人工智能作用于药物探索仍处于起步阶段,但是如果一旦实现将会让生物学释放出新的创新浪潮,并使21世纪真正成为属于它的纪元。
在这篇文章中,我们看了TypeDB是如何实现生物医学知识的符号化表示,以及如何改善ML来为药物探索做出贡献的。在药物探索中应用人工智能的科学家们使用TypeDB来分析疾病网络,更好地理解生物医学研究的复杂性,并发现新的和突破性的治疗方式。
崔皓,51CTO社区编辑,资深架构师,拥有18年的软件开发和架构经验,10年分布式架构经验。
原文标题:Artificial Intelligence in Drug Discovery,作者:Tomás Sabat
The above is the detailed content of Artificial Intelligence in Medical Discovery. For more information, please follow other related articles on the PHP Chinese website!
 Application of artificial intelligence in life
Application of artificial intelligence in life
 What is the basic concept of artificial intelligence
What is the basic concept of artificial intelligence
 What does bios mean?
What does bios mean?
 Two-way data binding principle
Two-way data binding principle
 What is LAN
What is LAN
 httpstatus500 error solution
httpstatus500 error solution
 linux view system information
linux view system information
 Solution to the problem that the input is not supported when the computer starts up
Solution to the problem that the input is not supported when the computer starts up




