

Author | Shanghai Jiao Tong University, Shanghai Artificial Intelligence Laboratory
Editor | ScienceAI
Recently, the joint team of Shanghai Jiao Tong University and Shanghai Artificial Intelligence Laboratory released a large 3D medical image segmentation model SAT (Segment Anything in radiology scans, driven by Text prompts), on 3D medical images (CT, MR, PET), based on text prompts to achieve universal segmentation of 497 types of organs/lesions in the human body. All data, codes, and models are open source.

Paper link:https://arxiv.org/abs/2312.17183
Code link:https://github.com/zhaoziheng/SAT
Data link:https://github .com/zhaoziheng/SAT-DS/
Research background
Medical image segmentation plays an important role in a series of clinical tasks such as diagnosis, surgical planning, and disease monitoring. However, traditional research trains "dedicated" models for each specific segmentation task, resulting in each "dedicated" model having a relatively limited scope of application and being unable to efficiently and conveniently meet a wide range of medical segmentation needs.
At the same time, large language models have recently achieved great success in the medical field, and to further promote the development of general medical artificial intelligence, it has become necessary to build a medical segmentation tool that can connect language and positioning capabilities.
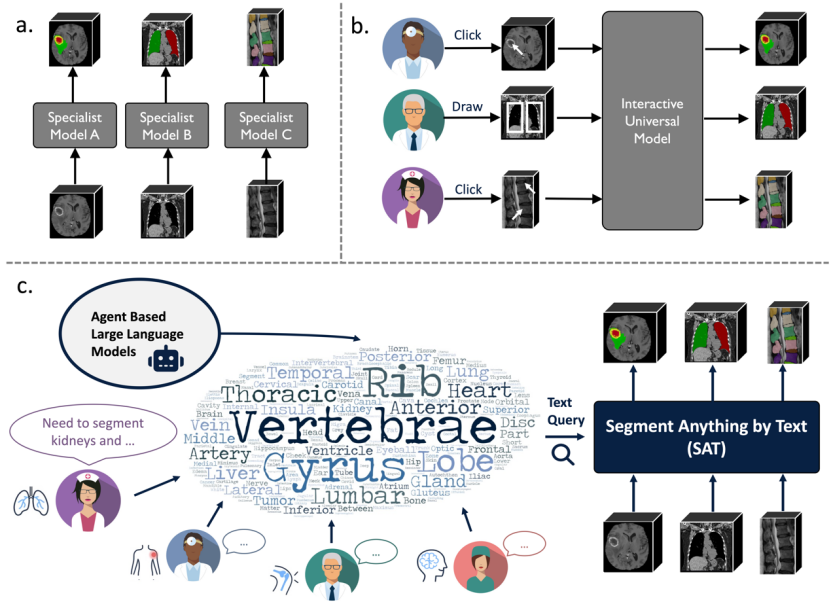
To overcome these challenges, researchers from Shanghai Jiao Tong University and Shanghai Artificial Intelligence Laboratory proposed the first general segmentation model for 3D medical images based on knowledge enhancement and using text prompts, called SAT (Segment Anything in radiology scans) , driven by Text prompts), and made the following three main contributions:
1. This study is the first to explore injecting human anatomy knowledge into a text encoder to accurately encode anatomical terms and achieve text prompts. A general medical segmentation model for radiology images.
2. This research builds the first multi-modal medical knowledge graph containing 6K+ human anatomy concepts. At the same time, the largest 3D medical image segmentation data set has been constructed, called SAT-DS, which brings together 72 public data sets, 22K+ images from CT, MR and PET modalities, and 302K+ segmentation annotations, covering the human body. 497 segmentation targets in 8 main parts.
3. Based on SAT-DS, this study trained two models of different sizes: SAT-Pro (447M parameters) and SAT-Nano (110M parameters), and designed experiments to verify the value of SAT from multiple angles: SAT The performance is equivalent to that of 72 nnU-Nets expert models (parameters are adjusted and optimized separately on each data set, a total of about 2.2B parameters), and shows stronger generalization ability on out-of-domain data; SAT can be used as a The basic segmentation model pre-trained on large-scale data can show better performance than nnU-Nets when transferred to specific tasks through downstream fine-tuning; in addition, compared with MedSAM based on box prompts, SAT can achieve more accurate and accurate performance based on text prompts. More efficient segmentation; finally, on clinical data outside the domain, the research team demonstrated that SAT can be used as a proxy tool for large language models, directly giving the latter the ability to localize and segment in tasks such as report generation.
The following will introduce the details of the original article from three aspects: data, model and experimental results.
Data construction
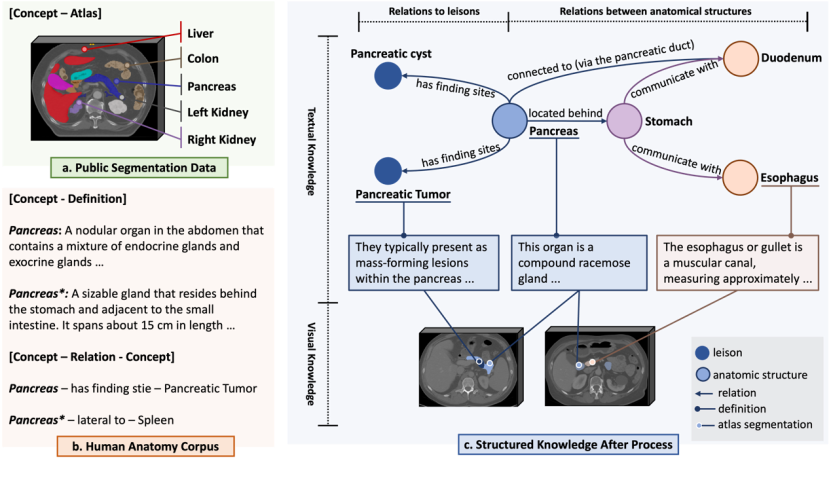
Multi-modal knowledge graph: In order to achieve accurate encoding of anatomical terms, the research team first collected a multi-modal knowledge graph containing 6K+ human anatomy concepts, the content of which comes from three sources :
1. Unified Medical Language System (UMLS) is a biomedical dictionary built by the U.S. National Library of Medicine. The research team extracted nearly 230K biomedical concepts and definitions, as well as a knowledge graph covering 1M+ mutual relationships.
2. Authoritative anatomy knowledge on the Internet. The research team screened 6,502 human anatomy concepts and retrieved relevant information from the Internet with the help of a retrieval-enhanced large language model, obtaining 6K+ concepts and definitions and a knowledge map covering 38K+ relationships between anatomical structures.
3. Public segmentation dataset. The research team collected a large-scale public 3D medical image segmentation data set and connected the segmented areas through anatomical concepts (category labels) with the knowledge in the above-mentioned text knowledge base to provide visual knowledge comparison.
SAT-DS:為了訓練通用分割模型,研究團隊建構了領域內最大規模的3D醫學影像分割資料集合SAT-DS。特別的,72個多樣的公開分割資料集被收集並整理,總計包括22186例3D影像,302033個分割標註,來自CT、MR和PET三種模態,以及涵蓋人體8個主要區域的497個分割類別(解剖學結構或病灶)。
為了盡可能降低異質資料集間的差異,研究團隊對不同資料集間的方向、體素間距、灰階值等影像屬性進行了標準化,用統一的解剖學術語系統命名了不同資料集中的分割類別。

圖 3:SAT-DS是一個大規模、多樣化的3D醫學影像分割資料集合,涵蓋人體8個主要區域共497個分割類別。
模型架構
知識注入:為了建構可以精確編碼解剖學術語的提示編碼器,研究團隊首先將多模態解剖學知識以對比學習的方式註入到文本編碼器。
如下圖a所示,用解剖學概念將多模態知識連接成對,隨後使用視覺編碼器(visual encoder)和文本編碼器(text encoder)分別編碼視覺和文本知識,透過對比學習在特徵空間中將解剖學結構的視覺特徵和文本知識對齊,並構建解剖學結構之間的關係,從而學習到對解剖學概念的更好編碼,作為提示引導視覺分割模型的訓練。
基於文字提示的通用分割:研究團隊進一步設計了基於文字提示的通用分割模型框架,如下圖b所示,包含文字編碼器、視覺編碼器、視覺解碼器與提示解碼器。
其中,考慮到同一解剖結構在不同影像中存在差異,提示解碼器(query decoder)使用視覺編碼器輸出的影像特徵增強解剖學概念特徵,即分割提示。最後,在分割提示與視覺解碼器輸出的像素級特徵間計算點積,得到分割預測結果。

模型測評
研究將SAT與兩個代表性的方法進行對比,即「專用」模型nnU-Nets和互動式通用分割模型MedSAM。測評包含了域內資料集測試(綜合分割效能)與zero-shot域外資料集測試(跨中心資料遷移能力)兩方面,評測結果從資料集、類別和人體區域三個層面進行了整合:
類別:不同資料集之間相同類別的分割結果進行匯總、平均;
區域:基於類別結果,將同一人體解剖區域內的類別結果進行匯總、平均;
資料集:傳統的分割模型評估方式,同一資料集內的分割結果進行平均;
與專用模型nnU-Nets的對比實驗
為了最大化nnU-Nets的性能,該研究在每個單獨的數據集上訓練nnU-Nets並與SAT對比,具體設定如下:
1. 在域內測試中,使用SAT-DS中的全部72個資料集進行測試和對比。對於SAT,使用72個訓練集的總和進行訓練,並在72個測試集上進行測試;對於nnU-Nets,匯總72個nnU-Nets在各自測試集上的結果作為一個整體。
2. 在域外測試中,進一步劃分72個資料集,使用其中的49個資料集(命名為SAT-DS-Nano)的訓練集訓練SAT-Nano,在10個域外的測試集上zero- shot測試;對於nnU-Nets,使用49個nnU-Nets在10個域外測試集上測試並彙總結果。

域內測試結果:從表1可以看到,SAT-Pro在域內測試中表現出與72個nnU-Nets十分接近的性能,並在多個區域上超越nnU-Nets。要注意的是,SAT可以只用一個模型完成72個分割任務,並且在模型尺寸上遠小於nnU-Nets的集合(如下圖c所示)。
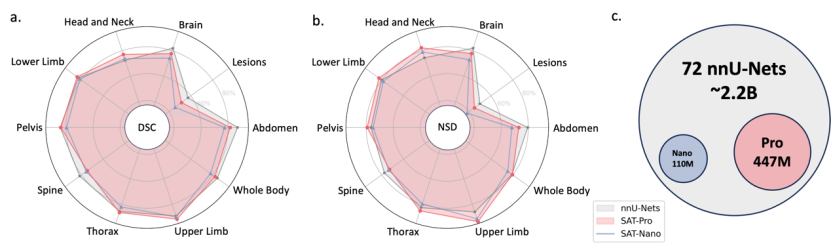
Fine-tuning migration test results: The study further tested SAT-Pro on each data set after fine-tuning separately, named SAT-Pro-Ft. As can be seen from Table 1, SAT-Pro-Ft has significant performance improvements in all areas compared to SAT-Pro, and exceeds nnU-Nets in overall performance.
Out-of-domain test results: As shown in Table 2, SAT-Nano surpassed nnU-Nets in 19 of the 20 indicators in 10 data sets, showing overall stronger migration capabilities.
Table 2: Comparison of out-of-domain tests between SAT-Nano, nnU-Nets, and MedSAM. The results are presented in units of data sets.

Comparative experiment with the interactive segmentation model MedSAM
This study directly uses MedSAM’s public checkpoint for testing and SAT comparison. The specific settings are as follows:
1. In the in-domain test, from 72 data We further screened 32 data sets used in MedSAM training for comparison.
2. In the out-of-domain test, 5 data sets that have not been used in MedSAM training were screened for comparison.
For MedSAM, consider two different Box prompts: using the smallest rectangle (Oracle Box) containing ground truth segmentation, recorded as MedSAM (Tight); adding random offsets based on the Oracle Box, recorded as MedSAM (Loose). At the same time, test the effect of Oracle Box directly as a prediction. For SAT, the model in the nnU-Nets comparison experiment is directly used to test on these data sets without retraining.
In-domain test results:As shown in Table 3, SAT-Pro performs better than MedSAM in almost all areas, and the overall performance of SAT-Pro and SAT-Nano is better than MedSAM. Although SAT-Pro performs worse than MedSAM on lesions, Oracle Box itself performs well enough on lesions as a prediction, even surpassing MedSAM on DSC. This indicates that the superior performance of MedSAM in segmenting lesions is likely to come from the strong prior information prompted by Box.
Table 3: In-domain test comparison of SAT-Pro, SAT-Nano and MedSAM, the results are integrated in units of regions or lesions.

Qualitative comparison: Figure 6 selects two typical examples from the results of the in-domain test for visual display to further compare SAT and MedSAM. As shown in Figure 6, in the segmentation of the myocardium, the Box prompt is difficult to distinguish between the myocardium and the ventricles wrapped by the myocardium, so MedSAM also mistakenly segmented the two together, which shows that the Box prompt is in a similar complex spatial relationship. It is easy to have ambiguities, leading to inaccurate segmentation.
In contrast, SAT based on text prompts (directly inputting the names of anatomical structures) can accurately distinguish between myocardium and ventricles. In addition, as can be seen in the intestinal tumor segmentation shown in Figure 6, Oracle Box is already a good prediction result for the lesion target, while the segmentation result of MedSAM may not be better than the obtained Box prompt.
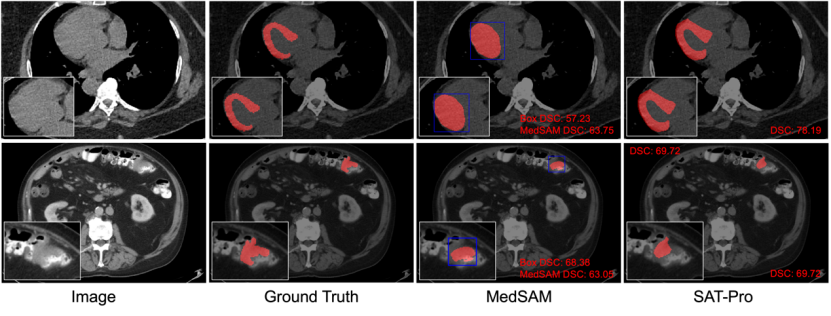
Figure 6: Qualitative comparison between SAT-Pro and MedSAM (Tight). Among them, MedSAM uses Oracle Box as a prompt, and the Box is marked in blue. The first row shows an example of myocardial segmentation; the second row shows an example of intestinal tumor segmentation.
Out-of-domain test results: As shown in Table 2, compared with MedSAM (Tight), SAT-Nano surpassed MedSAM in 5 out of 10 indicators in 5 data sets. MedSAM (Loose) has obvious performance degradation in all indicators, indicating that MedSAM is more sensitive to the offset of the Box prompt entered by the user.
Ablation experiment
When designing SAT, the visual backbone network and text encoder are two key parts. This research attempts to use different visual network structures or text encoders in the SAT framework, and general ablation experiments to explore their influence.
In order to save the cost of experiments, all SAT model training and testing in ablation experiments are performed on SAT-DS-Nano containing 49 datasets, which contains 13303 3D images, 151461 segmentation annotations, and 429 Split categories.
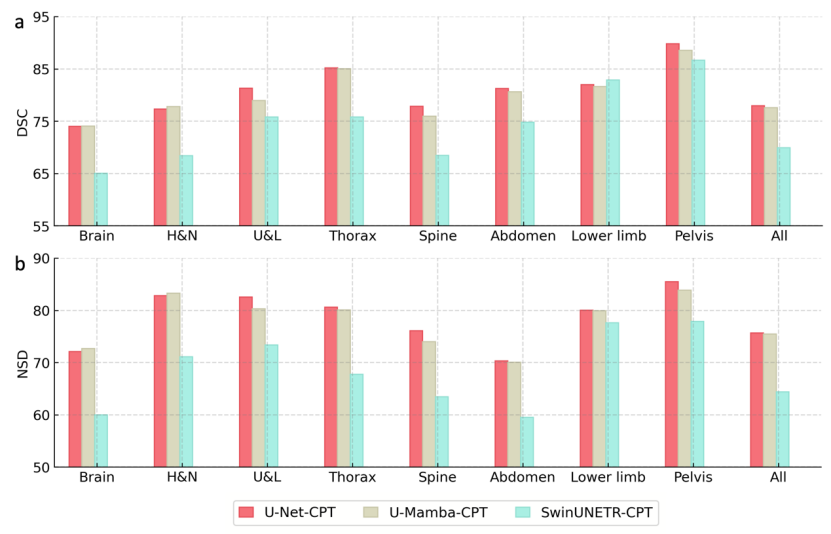
視覺骨幹網路:在SAT-Nano的框架下,研究選用了三種主流的分割網路結構進行對比,即U-Net(110M參數),SwinUNETR(107M參數)和U-Mamba(114M參數)。為了公平對比,在此消融實驗中控制它們的參數量大致接近。同時為了計算開銷,省略了知識注入的步驟,直接使用MedCPT(MedCPT是基於PubMed文獻,使用225M私有用戶點擊資料訓練的文本編碼器,在一系列醫療語言任務中取得了最好的表現)作為文本編碼器產生提示。三種變體分別記為U-Net-CPT、SwinUNETR-CPT和U-Mamba-CPT。
從圖7可以看到,使用U-Net與U-Mamba作為視覺骨幹網絡,最終的分割性能比較接近,其中U-Net略好於U-Mamba;而使用SwinUNETR時的分割性能有明顯下降。最後,研究團隊選擇U-Net作為SAT的視覺骨幹網路。
文本編碼器:在SAT-Nano的框架下,研究選擇了三種代表性的文本編碼器進行比較:使用上文提出的知識注入方式訓練的文本編碼器(記為Ours),使用先進的醫療文字編碼器MedCPT,使用沒有針對醫療資料微調的文字編碼器BERT-base。
為了公平,此消融實驗統一採用U-Net作為視覺網路。三種變體分別記為U-Net-Ours,U-Net-CPT和U-Net-BB。如圖8所示,總體而言,使用MedCPT相比使用BERT-base對於分割性能有微小的提升,表明領域知識對提供好的分割提示有一定的幫助;而使用該研究提出的文本編碼器在所有的類別上都取得了最好的性能,顯示建構多模態人體解剖學知識庫和知識注入對於分割模型有明顯的幫助。

長尾分佈是分割資料集的明顯特徵。如圖9中a圖和b圖所示,研究團隊研究了用於消融實驗的SAT-DS-Nano中429個類別的標註數量分佈。如果將標註數最多的10個類(前2.33%)定義為頭部,標註數最少的150個類別(後34.97%)定義為尾部類,可以發現尾部類的標註數量僅佔總體標註數的3.25 %。
研究進一步探討文本編碼器對長尾分佈中不同類別的分割結果影響。如圖9中c圖所示,研究團隊提出的編碼器在頭部、尾部和中間類別上都取得了最好的性能,其中尾部類別上的提升比頭部類更加明顯。同時,MedCPT在頭部類別上表現略低於BERT-base,但在尾部類別上效果更好。這些結果都表明,領域知識,特別是多模態人體解剖學知識的注入,對於長尾類別的分割有明顯幫助。

與大語言模型的結合
由於SAT可以基於文字提示進行分割,它可以被直接用作大語言模型的代理工具,提供分割能力。為了展示應用場景,研究團隊選擇了4個多樣的真實臨床數據,使用GPT4從報告中提取分割目標並調用SAT進行zero-shot分割,結果展示在圖10中。
可以看到,GPT-4可以很好地檢測到報告中重要的解剖學結構,並且調用SAT,在不需要任何數據fine-tune的情況下,在真實臨床圖像上很好地分割出這些目標。

研究價值
作為首個基於文字提示的3D醫療圖像通用分割大模型,SAT的價值體現在許多方面:
SAT建構了高效靈活的通用分割:SAT-Pro僅用一個模型,在廣泛的分割任務上展現出與72個nnU-Nets相當的性能,並且有更少的模型參數量。這顯示相較於傳統的醫療分割方法需要配置、訓練和部署一系列專用模型,SAT-Pro作為通用分割模型是更靈活高效的解決方法。同時,研究團隊也證明SAT-Pro在域外資料上泛化表現較好,能更好地滿足跨中心遷移等臨床需求。
SAT是基於大規模分割資料預訓練的基礎模型:SAT-Pro在大規模的分割資料集上進行訓練後,當透過微調遷移到特定資料集上時,表現出了明顯的效能提升,並比nnU-Nets整體性能更好。這顯示SAT可以被視為強大的基礎分割模型,可以透過微調遷移在特定任務上表現更好,從而平衡通用分割與專用分割的臨床需求。
SAT實現了基於文字提示的準確、魯棒分割:相比基於Box提示的交互式分割模型,SAT基於文字提示可以取得更準確和對提示魯棒的分割結果,並且可以節省使用者圈畫Box的大量時間,從而實現自動的、可批次化的通用分割。
SAT可以作為大語言模型的代理工具:研究團隊在真實臨床數據上展示了SAT可以和大語言模型無縫銜接,透過文本作為橋樑,直接為任何大語言模型提供分割和定位能力。這對進一步推動通用醫療智慧(Generalist Medical Artificial Intelligence)的發展有重要價值。
模型尺寸對於分割的影響:透過訓練兩種不同大小的模型:SAT-Nano和SAT-Pro,研究觀測到在域內測試中SAT-Pro相比SAT-Nano有明顯的提升。這暗示著在大規模資料集上訓練通用分割模型時,scaling-law依然適用。
領域知識對於分割的影響:研究團隊提出了第一個多模態的人體解剖學知識庫,並探索用知識增強提升通用分割模型的性能,特別是對長尾類別的分割。考慮到分割標註,特別是長尾類別上的標註,相對稀缺,這項探索對於建構通用的分割模型具有重要意義。
The above is the detailed content of The open source 3D medical large model SAT supports 497 organoids and has performance exceeding 72 nnU-Nets. It was released by the Shanghai Jiao Tong University team.. For more information, please follow other related articles on the PHP Chinese website!




