

Author | Cornell University Du Yuanqi
Editor | ScienceAI
As AI for Science receives more and more attention, people are more concerned about how AI can solve a series of scientific problems and can be successfully used for reference in other countries similar fields.
AI and small molecule drug discovery is one of the most representative and early explored fields. Molecular discovery is a very difficult combinatorial optimization problem (due to the discrete nature of the molecular structure) and the search space is very large and rugged. At the same time, it is very difficult to verify the properties of the searched molecules. It usually requires expensive experiments, at least simulation calculations, Quantum chemical methods to provide feedback.
With the rapid development of machine learning and thanks to early exploration (including the construction of simple and usable optimization goals and effect measurement methods), a large number of algorithms have been developed, including combinatorial optimization, search, sampling algorithms (genetic algorithms, Monte Carlo tree search, reinforcement learning, generative flow model/GFlowNet, Markov chain Monte Carlo, etc.), and continuous optimization algorithms, Bayesian optimization, gradient-based optimization, etc. At the same time, the existing relatively complete algorithm measurement benchmarks and relatively objective and fair comparison methods have also opened up a broad space for the development of machine learning algorithms.
Recently, researchers from Cornell University, University of Cambridge and Ecole Polytechnique Fédérale de Lausanne (EPFL) published a review article titled "Machine learning-aided generative molecular design" in "Nature Machine Intelligence".

Paper link:https://www.nature.com/articles/s42256-024-00843-5
This review reviews the application of machine learning in generative molecular design. Drug discovery and development requires optimizing molecules to meet specific physicochemical properties and biological activities. However, traditional methods are expensive and prone to failure due to the huge search space and discontinuous optimization functions. Machine learning accelerates the early-stage drug discovery process by combining molecule generation and screening steps.

Illustration: Generative ML assisted molecular design process.
Generative molecular design tasks
Generative molecular design can be divided into two major paradigms: distributed learning and goal-oriented generation, among which goal-oriented generation can be further divided into conditional generation and molecular optimization. The suitability of each method depends on the specific task and the data involved.
Distribution learning (distribution learning)
Conditional generation (conditional generation)
Molecular optimization plays a key role in drug discovery by refining the properties of drug candidates to improve their safety, efficacy and pharmacokinetic properties. Involves making small modifications to candidate molecular structures to optimize drug properties such as solubility, bioavailability, and target affinity, thereby improving therapeutic potential and increasing success with clinical endpoints.
Illustrations: Illustrations of generation tasks, generation strategies, and molecular characterization.
Molecular generation process
Molecular generation is a complex process including many different combination units. We list the representative work in the figure below and introduce the representative units of each part.
Molecular Representation
When developing molecularly generated neural architectures, it is first necessary to determine machine-readable input and output representations of the molecular structure. The input representation helps inject appropriate inductive biases into the model, while the output representation determines the optimized search space for the molecule. The representation type determines the applicability of the generation method, for example, discrete search algorithms can only be applied to combinatorial representations such as graphs and strings.
While various input representations have been studied, the trade-offs between representation types and the neural architectures that encode them are not yet clear. Representation transformations between molecules are not necessarily bijective; for example, density maps and fingerprints cannot uniquely identify molecules, and further techniques are needed to solve this non-trivial mapping problem. Common molecular representations include strings, two-dimensional topological graphs, and three-dimensional geometric graphs.
Representation granularity is another consideration in generative model design. Typically, methods utilize atoms or molecular fragments as basic building blocks during generation. Fragment-based representation refines molecular structures into larger units containing groups of atoms, carrying hierarchical information such as functional group identification, thereby aligning with traditional fragment-based or pharmacophore drug design approaches.
Generative methods
Deep generative models are a class of methods that estimate the probability distribution of data and sample from a learning distribution (also called distribution learning). These include variational autoencoders, generative adversarial networks, normalizing flows, autoregressive models, and diffusion models. Each of these generation methods has its use cases, pros and cons, and the choice depends on the required task and data characteristics.
Generation strategy
Generation strategy refers to the way the model outputs the molecular structure, which can generally be divided into one-time generation, sequential generation or iterative improvement.
One-shot generation:One-shot generation generates the complete molecular structure in a single forward pass of the model. This approach often struggles to generate realistic and reasonable molecular structures with high accuracy. Furthermore, one-shot generation often cannot satisfy explicit constraints, such as valence constraints, which are crucial to ensure the accuracy and validity of the generated structure.
Sequential Generation:Sequential generation builds a molecular structure through a series of steps, usually by atoms or fragments. Valence constraints can be easily injected into sequential generation, thereby improving the quality of the generated molecules. However, the main limitation of sequential generation is that the order of generated trajectories needs to be defined during training and is slower in inference.
Iterative improvement:Iterative improvement adjusts the prediction by predicting a series of updates, circumventing the difficulties in one-shot generation methods. For example, the cyclic structure module in AlphaFold2 successfully refined the backbone framework, an approach that inspired related molecule generation strategies. Diffusion modeling is a common technique that generates new data through a series of noise reduction steps. Currently, diffusion models have been applied to a variety of molecule generation problems, including conformational generation, structure-based drug design, and linker design.
Optimization strategy
Combination optimization:For the combinatorial encoding of molecules (pictures or strings), techniques in the field of combinatorial optimization can be directly applied.
Continuous Optimization:Molecules can be represented or encoded in continuous domains, such as point clouds and geometric maps in Euclidean space, or deep generative models that encode discrete data in continuous latent space.

Evaluation of Generative Machine Learning Models
Evaluating generative models requires computational evaluation and experimental verification. Standard metrics include effectiveness, uniqueness, novelty, etc. Multiple metrics should be considered when evaluating a model to fully assess build performance.
Experimental verification
產生的分子必須透過濕法實驗來進行明確的驗證,這與現有研究主要關注計算貢獻形成鮮明對比。雖然生成模型並非沒有弱點,但預測與實驗之間的脫節也歸因於進行此類驗證所需的專業知識、昂貴的費用、以及漫長的測試週期。

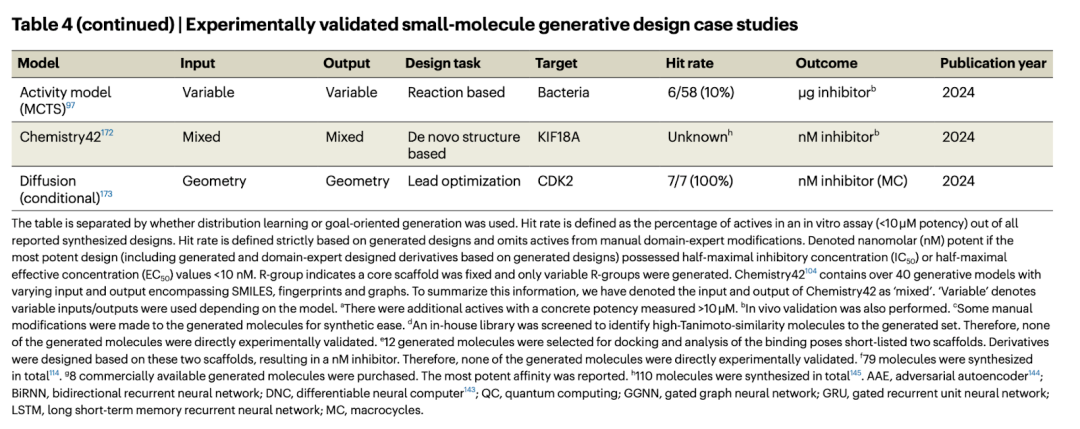
產生模型規律
大多數報告實驗驗證的研究使用 RNN 和/或 VAE,並以 SMILES 作為操作對象。我們總結了四個主要觀察點:
未來方向
儘管機器學習演算法為小分子藥物發現帶來了曙光,但是還有更多的挑戰與機遇需要面對。
挑戰
機會
作者: 杜沅岂,康奈尔大学计算机系二年级博士生,主要研究兴趣,几何深度学习,概率模型,采样,搜索,优化问题,可解释性,与在分子探索领域的应用,具体信息见:https://yuanqidu.github.io/。
The above is the detailed content of 'Encyclopedia' of AI small molecule drug discovery, reviewed by researchers from Cornell, Cambridge, EPFL and others published in Nature sub-journal. For more information, please follow other related articles on the PHP Chinese website!
 oracle clear table data
oracle clear table data What to do if notepad.exe is not responding
What to do if notepad.exe is not responding createprocess failure reason
createprocess failure reason Tutorial on buying and selling Bitcoin on Huobi.com
Tutorial on buying and selling Bitcoin on Huobi.com There are several ways to position CSS position
There are several ways to position CSS position How to turn on and off Douyin Xiaohuoren
How to turn on and off Douyin Xiaohuoren What should I do if my QQ account is stolen?
What should I do if my QQ account is stolen? How to optimize a single page
How to optimize a single page



